Abstract
This study illustrates that Plekhm1 is an essential protein for bone resorption, as loss-of-function mutations were found to underlie the osteopetrotic phenotype of the incisors absent rat as well as an intermediate type of human osteopetrosis. Electron and confocal microscopic analysis demonstrated that monocytes from a patient homozygous for the mutation differentiated into osteoclasts normally, but when cultured on dentine discs, the osteoclasts failed to form ruffled borders and showed little evidence of bone resorption. The presence of both RUN and pleckstrin homology domains suggests that Plekhm1 may be linked to small GTPase signaling. We found that Plekhm1 colocalized with Rab7 to late endosomal/lysosomal vesicles in HEK293 and osteoclast-like cells, an effect that was dependent on the prenylation of Rab7. In conclusion, we believe PLEKHM1 to be a novel gene implicated in the development of osteopetrosis, with a putative critical function in vesicular transport in the osteoclast.
Introduction
Osteopetrosis is a genetically and clinically heterogeneous bone disorder characterized by a reduction in bone resorption and a generalized net accumulation of skeletal mass. In humans, several subforms are classified according to severity, mode of inheritance, and age of onset (reviewed in ref. 1). Not all the genetic mutations that lead to human osteopetrosis are known yet. The causative genes identified so far all play a role in acidification of the resorption lacuna, and loss-of-function mutations in these genes severely affect mature osteoclast function. The CA2 gene produces the protons necessary for acidification of the resorption lacuna, the extracellular compartment between the bone tissue and the osteoclast where bone resorption occurs (2). The α3 subunit of the H+ ATPase, encoded by the TCIRG1 gene, is involved in the transportation of these protons through the ruffled border into the resorption lacuna (3, 4), while chloride channel 7 (CLC7), encoded by the CLCN7 gene, translocates chloride ions to maintain electroneutrality (5–7). Finally, the OSTM1 gene encodes a type I transmembrane protein with E3 ubiquitin ligase activity (8–11). The exact function of Ostm1 remains elusive, but a recent report identified the Ostm1 protein as a β subunit of CLC7 and demonstrated that it requires CLC7 to localize to lysosomes (12).
In animals, mutations affecting both formation and function of osteoclasts have been described (reviewed in ref. 13). The osteopetrotic mutations in animals, both spontaneous and induced, are all inherited in an autosomal-recessive manner. Extensive variations in severity, life expectancy, and osteoclast features are observed, and sometimes tissues other than bone are also affected. One of the spontaneous mutations is the incisors absent (ia) rat, first described in 1941 (14), which exhibits a generalized skeletal sclerosis and delay of tooth eruption. Histologic analysis shows that ia mutants have 2 to 3 times more osteoclasts than do normal littermates and that these cells exhibit an extended clear zone, an actin-rich area that facilitates tight attachment to the extracellular matrix (15). Furthermore, ia osteoclasts lack ruffled borders but contain numerous small cytoplasmic vesicles. The absence of extracellular tartrate-resistant acid phosphatase (TRAP), concomitant with accumulation of the enzyme in numerous intracellular vesicles, strongly suggests a dysfunction of the secretory pathways (15). Although a spontaneous partial recovery of the osteopetrotic phenotype is observed 30 to 50 days after birth (16), the defect is more rapidly reversed by transplantation of hematopoietic stem cells from normal littermates in irradiated ia rats. Osteoclasts with ruffled borders that are indistinguishable from those in normal littermates then appear, and the dense sclerotic skeleton is rapidly remodeled and becomes normal. This indicates that the primary defect is intrinsic to the osteoclast (17–19).
Although ia rats have been well studied phenotypically, the underlying genetic defect has remained unknown. In order to elucidate the gene responsible, we previously performed segregation analysis that delineated a 4.7-cM region on rat chromosome 10q32.1 in which the disease-causing gene is located (20). In the present study, we identified this gene as pleckstrin homology domain–containing family M (with RUN domain) member 1 (PLEKHM1), identified a mutation in this gene as a cause of osteopetrosis in humans, and provided some insight into the function of the Plekhm1 protein in bone resorption.
Results
Refinement of the ia candidate region.
After our initial gene localization to a 4.7-cM region on rat chromosome 10q32.1, genetic analysis of extra markers and 50 additional mutant animals (40 F4, 9 F6, and 1 F8) allowed us to narrow the ia candidate region to 2.2 cM, flanked by the markers D10Rat205 and D10Got137 (data not shown).
Identification of the ia mutation.
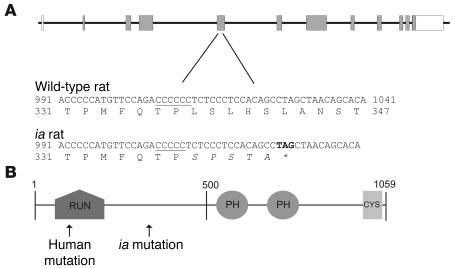
We performed sequencing analysis of several genes located in this interval on kidney cDNA from normal and ia rats and identified in the latter a homozygous deletion of 1 cytosine in the fourth coding exon of the PLEKHM1 gene. This deletion — located in a stretch of 6 cytosines on cDNA position 1,007–1,012, with position 1 at the start codon — causes a frameshift mutation after codon 337, followed by 5 additional unrelated amino acids and a stop codon, thus yielding a truncated protein (Figure 1A). Homozygosity for this deletion was found in all 78 mutant animals.
Figure 1. The Plekhm1 gene and protein.
(A) Genomic structure of the rat plekhm1 gene and the ia mutation. The plekhm1 gene consists of 12 exons (boxes) with the start codon located in exon 2. In ia rats, 1 cytosine (located in a stretch of 6 cytosines, underlined) is deleted in exon 5 of the plekhm1 gene. This results in a frameshift mutation followed by 5 divergent amino acids and a stop codon (asterisk). (B) Domain structure of the Plekhm1 protein. The Plekhm1 protein consists of a RUN domain, 2 PH domains, and a cysteine-rich (CYS) domain. The positions of the ia and human mutations are indicated.
The PLEKHM1 gene.
The PLEKHM1 gene consists of 12 exons with the start codon located in exon 2, has a transcript length of 5,262 bp, and comprises a genomic region of 54.8 kb. It has orthologs in several organisms. Conservation of PLEKHM1 is strong among mammalian species, with mouse and human being 93% and 83% identical, respectively, to rat. The rat amino acid sequence is conserved for 82.3% and 93.5% compared with the human and mouse Plekhm1 proteins, respectively. We found 1 transcribed pseudogene located at a distance of approximately 2 Mb from the PLEKHM1 gene in humans only. However, sequence analysis of the pseudogene cDNA revealed numerous nucleotide substitutions, partial deletion of exon 3 including part of the RUN domain, and deletion of exon 4, which resulted in a translational frameshift and subsequently a premature stop codon. This 261–amino acid translation product of the pseudogene, if stable, probably has no function or an altered function compared with the full-length 1,057–amino acid Plekhm1 translation product.
Expression pattern of Plekhm1.
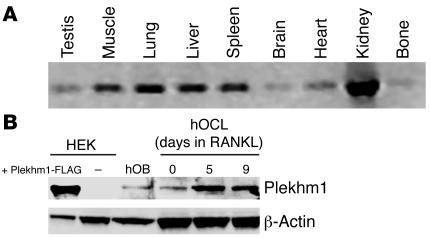
To reveal the expression pattern of the plekhm1 gene, a panel of rat tissue cDNAs was amplified using primers that overlap the entire coding sequence. The rat plekhm1 gene was expressed in all tissues tested (testis, skeletal muscle, lung, liver, spleen, brain, heart, kidney, and bone), and no alternative splice variants were detected (Figure 2A). We also examined by RT-PCR and sequence analysis the expression of the human PLEKHM1 gene and its transcribed pseudogene in several tissues, including cultures of differentiated osteoclasts, and found that both transcripts were expressed in all tissues examined (data not shown). Western blot analysis demonstrated that the Plekhm1 protein was expressed at relatively low levels in human monocytes. However, expression of Plekhm1 increased during RANKL-induced differentiation of both human M-CSF–dependent monocytes (Figure 2B; 2.9-fold increase as assessed by densitometry; n = 4) and RAW 264 macrophage-like cells (data not shown) into osteoclast-like cells. Expression of Plekhm1 in HEK cells was barely detectable, while expression in human osteoblasts was much less than in either human monocytes or osteoclast-like cells (6.1-fold less than in osteoclast-like cells as assessed by densitometry; n = 4). Although the antibody cross-reacted with a number of other proteins, Plekhm1 was identified on the blots by using HEK293 cells transfected with Plekhm1-FLAG as a positive control, which demonstrated that the Plekhm1 protein had a molecular mass of 140 kDa (Figure 2B).
Figure 2. Expression pattern of Plekhm1.
(A) The plekhm1 gene was expressed in all tissues examined, as demonstrated by a rat cDNA multiple tissue panel. (B) Western blot results of the Plekhm1 protein (confirmed by subsequent incubation of the same blot with the anti-FLAG antibody; data not shown) demonstrated that the Plekhm1 protein was expressed in human monocytes (hOCL) and osteoblasts (hOB). Moreover, expression of Plekhm1 increased during differentiation of human monocytes into osteoclasts by treatment with RANKL.
Screening for mutations in the human PLEKHM1 gene.
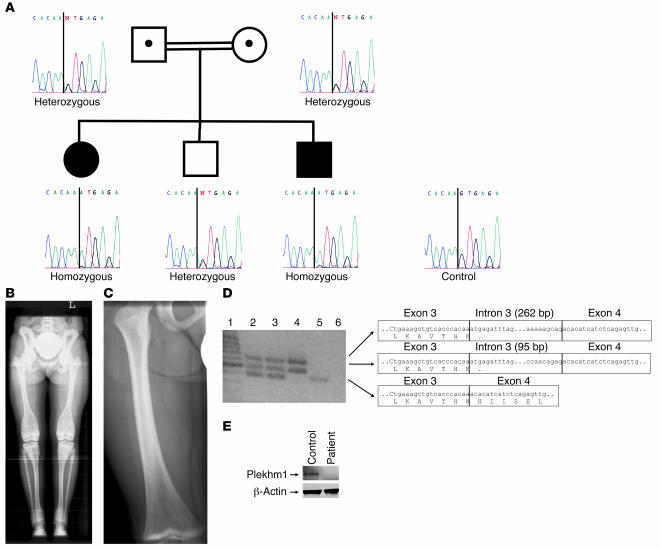
Based on the data obtained from the ia rat, we considered PLEKHM1 as a candidate gene for human osteopetrosis and performed mutation analysis of the entire coding sequence of the PLEKHM1 gene in 43 patients diagnosed with various forms of osteopetrosis. While most were excluded for mutations in the other known osteopetrosis genes, we identified a patient with a homozygous G→A transition at position +1 of the donor splice site of intron 3 (Figure 3A). She was diagnosed with an autosomal-recessive intermediate form of the disease. When the patient was 7 years old, an “Erlenmeyer flask” deformity of the distal femora was detected upon radiographic examination (Figure 3B). At the age of 14 years, she suffered from pain in the left leg during walking, and a chondrolysis of the left hip was observed. No other clinical symptoms were reported. As Plekhm1 is highly expressed in the kidney, we measured the patient’s creatinine clearance and found it to be 130 mg/ml, indicative of normal renal function. Her parents, carriers of the mutation, were related to each other and were clinically normal. The oldest brother was heterozygous for the mutation and was clinically and radiologically normal. The youngest brother was homozygous for the mutation but had not yet developed clinical symptoms. However, radiologic examination at the age of 8 months and again at 2 years did show the presence of dense metaphyseal bands (Figure 3C), which have also been reported in a case of decreased bone resorption due to lead poisoning (21). The mutation was not found in any of the 100 control chromosomes that we examined. To study the effect of the mutation at the RNA level, an RT-PCR assay spanning exons 3–4 was designed, amplifying only the PLEKHM1 gene and not the pseudogene. RT-PCR on total RNA isolated from cultured lymphoblasts resulted in a 382-bp product in an unrelated control, but not in the osteopetrotic patient. In fact, 2 larger products were present (Figure 3D). Direct sequencing revealed that the 2 forms contained 95 bp or 262 bp of intron 3 due to the presence of cryptic splice donor sites in intron 3. Both alternative splicing products led to a premature stop codon and a highly truncated gene product (99 amino acids). As expected, RT-PCR on total RNA isolated from the heterozygous carriers resulted in 3 fragments, corresponding to the 1 normal and 2 aberrant transcripts. Western blot analysis demonstrated that the Plekhm1 protein was not present in lysates of osteoclasts generated from PBMCs from the affected patient, in contrast to lysates of osteoclasts generated from monocytes of an unaffected individual (Figure 3E).
Figure 3. Mutation analysis of a family with autosomal-recessive osteopetrosis.
(A) Sequencing analysis showed a homozygous G→A transition at position +1 of intron 3 of the PLEKHM1 gene in the patient and the youngest brother. (B) Full-leg radiograph from the affected patient: cortical sclerosis of the pelvic bones, particularly at the iliac wings. Note the band-like sclerosis of the vertebral endplates (rugger jersey spine) and the inhomogeneous sclerosis at the metadiaphyses of the distal femora, tibiae and fibulae, and proximal fibulae and tibiae. Also note the broadening of the involved segments of the long bones (“Erlenmeyer flask” deformity). (C) Radiograph of the right femur of the youngest brother at 2 years of age showing the presence of a dense metaphyseal band at the distal metaphysis. (D) RT-PCR amplification across exons 3–4 of the PLEKHM1 gene. The sequence of the corresponding PCR products is presented. Lane 1, size marker; lane 2, father; lane 3, mother; lane 4, affected patient; lane 5, unrelated control; lane 6, blank. (E) Western blot analysis demonstrated that the Plekhm1 protein was not present in osteoclast lysates from the affected patient, in contrast to lysates from a healthy individual.
Defective bone resorption in osteoclasts from the patient with the PLEKHM1 mutation.
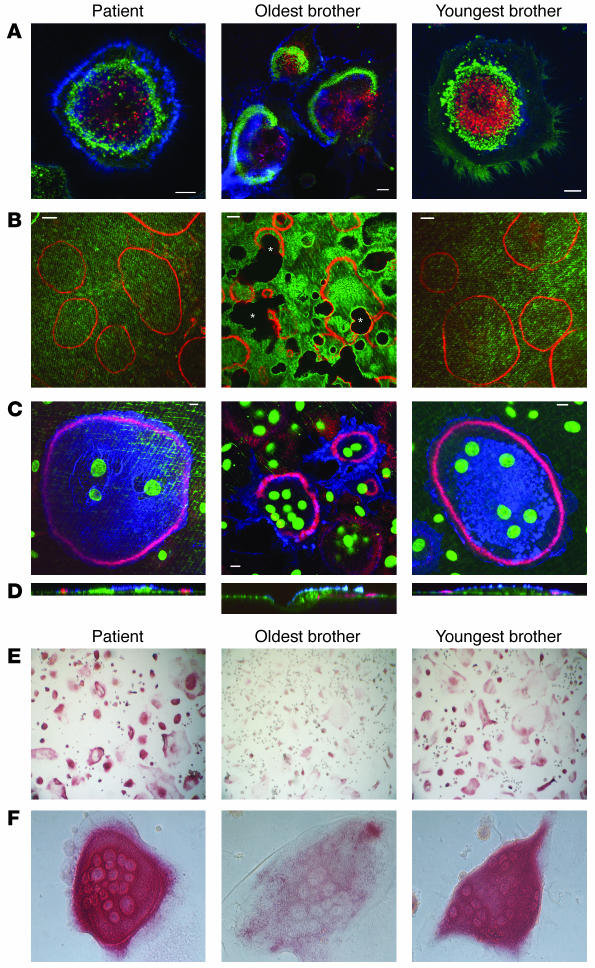
We generated osteoclasts from the patient and her 2 brothers on dentine discs by culturing PBMCs with M-CSF and RANKL. Osteoclasts formed normally from all 3 siblings, as demonstrated by the appearance of large, multinucleated, vitronectin receptor–positive (VNR-positive) cells. Osteoclasts from the patient and the youngest brother, also homozygous for the PLEKHM1 mutation, adhered to dentine by a ring of punctate, F-actin–rich podosomes, a characteristic feature of mature osteoclasts. However, these podosomes were arranged into peripheral belts (Figure 4, A and C) characteristic of nonresorbing osteoclasts and did not mature into the denser, more centrally localized F-actin rings characteristic of actively resorbing osteoclasts (22) seen in the cells from the unaffected older brother, who was heterozygous for the mutation (Figure 4A). Accordingly, osteoclasts from the patient and youngest brother showed hardly any evidence of resorptive activity when cultured on dentine discs, whereas osteoclasts from the older brother resorbed large areas of the dentine (Figure 4B). In addition, the osteoclasts derived from the patient and the youngest brother were often larger, despite generally having fewer nuclei, and had a more flattened morphology than did the active osteoclasts from the unaffected older brother (Figure 4, C and D). Using the acidotropic probe lysotracker, we also found that osteoclasts from the patient and youngest brother generated acidic vesicles, indicating that the formation of these vesicles was not defective (Figure 4A). Moreover, TRAP, as detected in vesicular structures by histochemical staining, appeared much more abundant in osteoclasts from the patient and the youngest brother than in those from the older brother (Figure 4, E and F).
Figure 4. Characterization of the osteoclasts from the family with autosomal-recessive osteopetrosis.
Cells were cultured on dentine discs (A–D) or plastic (E and F) in the presence of RANKL for 10 days and then fixed, stained, and analyzed by confocal microscopy (A–D) or conventional microscopy (E and F). (A) Staining of F-actin with FITC-phalloidin (green stain), acidic vesicles with lysotracker (red stain), and osteoclast membrane with anti-VNR antibodies (blue stain). (B) Staining of the dentine surface with FL-ALN (green stain) and F-actin with TRITC-phalloidin (red stain). Dark areas correspond to resorption pits (asterisks). (C) Staining of the dentine surface with FL-ALN and nuclei with Sytox Green (green); F-actin with TRITC-phalloidin (red); osteoclast membrane with anti-VNR antibodies (blue). A–C represent 1-μm xy optical sections; D represents zx reconstructions of osteoclasts in C. Scale bar: 10 μm. (E and F) Staining for TRAP in osteoclasts. Original magnification, ×10 (E) and ×40 (F).
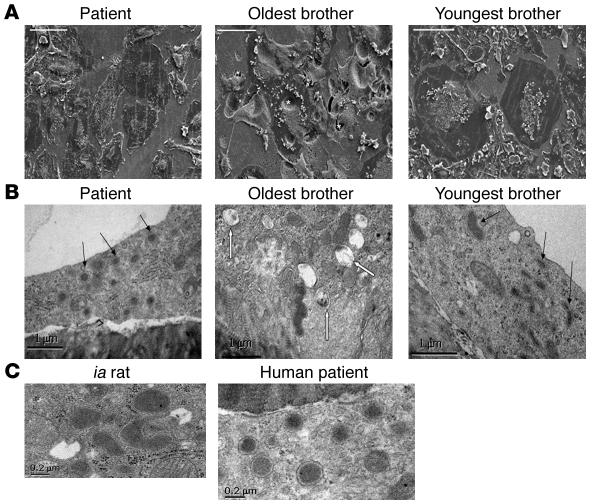
Scanning EM demonstrated osteoclasts of normal size and resorptive activity in cultures from the oldest brother. In accordance with the confocal microscopy data, osteoclasts from both the patient and the youngest brother were larger and more flattened, and showed little evidence of resorptive activity (Figure 5A). Transmission EM demonstrated that the cytoplasm of these nonresorbing osteoclasts contained numerous vacuoles with electron-dense material (Figure 5B) that were identical in appearance to the vesicles found in the ia rat (Figure 5C), which have previously been shown to contain TRAP enzyme by immunoelectron microscopy (15). These vesicles were rarely seen in resorbing osteoclasts, which instead contained abundant multivesicular bodies, structures previously associated with transcytosis of resorbed material by the cell (S. Nesbitt, unpublished observations).
Figure 5. Electron microscopy of osteoclasts generated from the family with autosomal-recessive osteopetrosis.
(A and B) Osteoclasts were formed from all 3 siblings, then fixed and analyzed by scanning EM or transmission EM. (A) Scanning EM images show that the oldest brother produced normal, actively resorbing osteoclasts excavating deep resorption pits (denoted by asterisks), whereas both the patient and the youngest brother formed large, very flat osteoclasts with little evidence of resorption. (B) Transmission EM demonstrated that both the patient and the youngest brother produced osteoclasts containing large numbers of electron-dense granules (black arrows), whereas the resorbing osteoclasts formed from the oldest brother contained many multivesicular bodies (white arrows). (C) The electron-dense vesicles were identical in ultrastructure to those seen in the ia rat, which are known to contain TRAP. Scale bars: 100 μm (A), 1 μm (B), 0.2 μm (C).
Functional domains of the Plekhm1 protein.
To date, part of the Plekhm1 protein has been described as the AP162 adaptor protein, without indication of a role in bone metabolism (23). In order to determine its potential function, we first analyzed the rat Plekhm1 protein sequence in silico. Analysis with the Signal P3.0 (24) and TMHMM3.0 programs (25) predicted that the Plekhm1 protein is a nonsecretory protein without signal peptide and does not contain any transmembrane helices. The InterPro database (26) predicted 4 functional domains: a RUN domain, 2 pleckstrin homology (PH) domains, and a cysteine-rich domain (Figure 1B). The latter (amino acids 990–1,043 of the Plekhm1 protein) is predicted to be either a RING finger domain involved in the ubiquitination pathway (27) or a protein kinase C–conserved region 1 domain essential for diacylglycerol/phorbol ester binding (28). RUN domains (amino acids 41–182 of the Plekhm1 protein) encode a globular structure consisting of 6 conserved blocks. They are found in a number of proteins, several of which have been demonstrated to interact with small GTPases (29–34). The PH domain (amino acids 537–628 and 686–781 of the Plekhm1 protein) consists of about 100 residues and can be divided into 6 subdomains. PH domains are found in a wide variety of proteins involved in intracellular signaling (including several that interact with small GTPases) or as constituents of the cytoskeleton targeting proteins to the cytosolic face of cellular membranes (35–40).
Plekhm1 localizes with Rab7 on late endosomes/lysosomes.
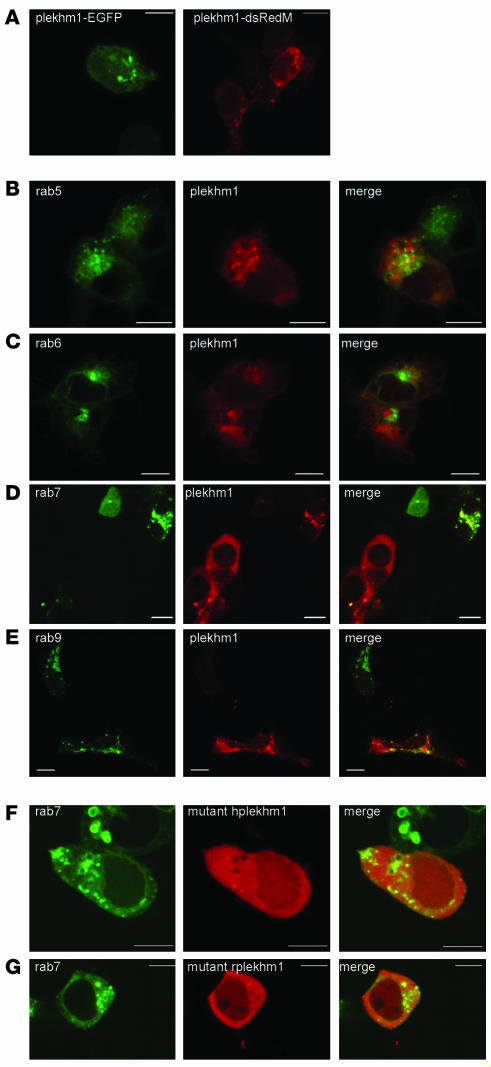
To determine the subcellular localization of the Plekhm1 protein, we transfected HEK293 cells with a Plekhm1–enhanced GFP (Plekhm1-EGFP) or Plekhm1-dsRedMonomer (Plekhm1-dsRedM) construct. Plekhm1 showed a diffuse cytoplasmic distribution, but was also partially associated with the membranes of intracellular vesicles that were often abnormally enlarged in transiently transfected cells (Figure 6A and data not shown). Interestingly, overexpression of some endosomal/lysosomal Rab GTPases, such as Rab5, Rab7, and Rab9, also caused abnormal enlargement of endosomes, likely the result of strong stimulation of intracellular transport (41–43), which suggests that Plekhm1 may be a component of Rab-mediated vesicular transport. This possibility is supported by the presence of a RUN domain within Plekhm1, a domain that has been identified in some proteins known to interact with Rab GTPases (29). We therefore examined whether overexpressed Plekhm1 colocalizes with Rab GTPase proteins, which are localized to specific vesicular compartments within the cell. Rab5 is localized to early endosomes (44), Rab6 to the Golgi apparatus (45), Rab7 to the late endosomal/lysosomal compartment (46–48), and Rab9 to late endosomes (49). Using overexpression studies, we found that Plekhm1-dsRedM showed no colocalization with EGFP-Rab5 or EGFP-Rab6 (Figure 6, B and C), partially colocalized with EGFP-Rab9 (Figure 6E), and completely colocalized with EGFP-Rab7 (Figure 6D and Figure 7A), indicating that the Plekhm1 protein is localized to late endosomes/lysosomes. Interestingly, coexpression of EGFP-Rab7 and Plekhm1-dsRedM also resulted in a striking recruitment of the Plekhm1 protein to Rab7-positive vesicles and a concomitant complete loss of cytoplasmic Plekhm1-dsRedM (Figure 6D), an effect that was not seen with overexpression of EGFP-Rab9 and Plekhm1-dsRedM (Figure 6E). In addition, EGFP-Rab7 showed slightly increased association with vesicles and reduced cytosolic localization in cells that also expressed Plekhm1 (Figure 6D). Identical results were found with a Plekhm1-FLAG construct (data not shown), demonstrating that the dsRedM tag did not influence the localization of Plekhm1. To determine the localization of the mutated Plekhm1 proteins, we transfected HEK293 cells with truncated human or rat Plekhm1-dsRedM constructs corresponding to the mutations found in the osteopetrotic patient and the ia rat. Both mutant Plekhm1 proteins showed a diffuse cytoplasmic distribution and did not colocalize with Rab7 to the endosomal vesicles (Figure 6, F and G).
Figure 6. Subcellular localization of Plekhm1 in HEK 293 cells.
HEK293 cells were transfected with plasmids, then fixed 24 hours later and analyzed by confocal microscopy. (A) Both Plekhm1-EGFP and Plekhm1-dsRedM displayed a diffuse cytoplasmic distribution, but were also clearly associated with intracellular vesicles, which frequently became abnormally enlarged. (B–E) HEK293 cells were cotransfected with Plekhm1-dsRedM and EGFP-Rab5 (B), EGFP-Rab6 (C), EGFP-Rab7 (D), or EGFP-Rab9 (E). Plekhm1 did not colocalize with either Rab5 or Rab6, but partially colocalized with Rab9 and completely colocalized to intracellular vesicles with Rab7. Interestingly, Plekhm1 showed more pronounced localization to these Rab7-positive vesicles when cotransfected with EGFP-Rab7. (F and G) HEK293 cells were transfected with EGFP-Rab7 and Plekhm1-dsRedM mutants corresponding to the mutations found in the osteopetrotic patient (hplekhm1; F) and the ia rat (rplekhm1; G). Neither mutant form of Plekhm1 colocalized with Rab7. Panels represent 1-μm xy optical sections. Scale bars: 10 μm.
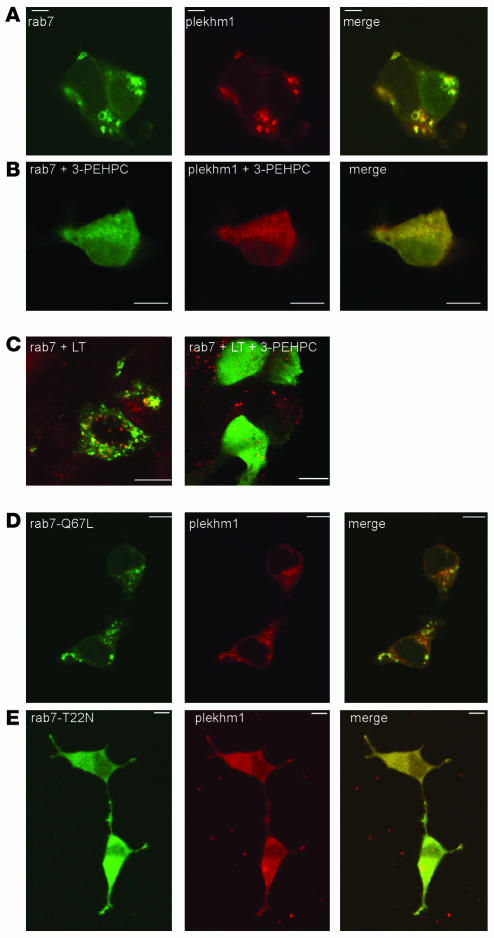
Figure 7. Localization of Plekhm1 to endosomes is dependent on prenylated, active Rab7.
HEK293 cells were incubated for 24 hours in the presence of the Rab GGTase inhibitor 3-PEHPC (1 mM) immediately following transfection with EGFP-Rab7 and Plekhm1-dsRedM. (A and B) The vesicular localization of both Rab7 and Plekhm1 (A) was disrupted by 3-PEHPC (B). (C) Acidic vesicles were stained with lysotracker (LT) prior to fixation. No effect of 3-PEHPC on the integrity of the endosomal/lysosomal compartment was observed. (D and E) HEK293 cells were transfected with Plekhm1-dsRedM and constitutively active EGFP–Rab7-Q67L (D) or dominant-negative EGFP–Rab7-T22N (E). Plekhm1 and Rab7-Q67L colocalized on intracellular vesicles, whereas Rab7-T22N and plekhm1 were both diffusely distributed throughout the cytoplasm. Panels represent 1-μm xy optical sections. Scale bars: 10 μm.
Because association of Rabs with cell membranes is dependent on posttranslational geranylgeranylation at the C terminus by Rab geranylgeranyl transferase (GGTase; ref. 50), we studied the subcellular distribution of Plekhm1-dsRedM and EGFP-Rab7 in HEK293 cells following treatment with the specific Rab GGTase inhibitor 3-PEHPC (51). No apparent alteration of the lysosomal compartment in HEK293 cells was observed after treatment with 3-PEHPC, as indicated by staining with the acidotropic dye lysotracker (Figure 7C). However, treatment with 3-PEHPC completely disrupted the endosomal localization of not only EGFP-Rab7, but also Plekhm1-dsRedM, which both became diffusely distributed throughout the cytosol (Figure 7, A and B). These results further indicate that the localization of Plekhm1 to late endosomes/lysosomes is dependent on Rab7.
We also coexpressed Plekhm1-dsRedM with dominant-negative or constitutively active EGFP-Rab7 mutants (the subcellular distributions of which have been described previously; refs. 41, 52) and analyzed the subcellular localization of the Plekhm1 protein. When the GTP-bound form of Rab7 (EGFP–Rab7-Q67L) and Plekhm1 were coexpressed in HEK293 cells, Plekhm1 was largely colocalized with Rab7-Q67L on vesicular structures (Figure 7D), similar to the results with wild-type Rab7. However, coexpression of the GDP-bound form of Rab7 (EGFP–Rab7-T22N) and Plekhm1 resulted in a mainly cytosolic, diffuse distribution of both Rab7-T22N and Plekhm1 (Figure 7E). These data are consistent with a role for Rab7 in the localization of Plekhm1 to late endosomes/lysosomes.
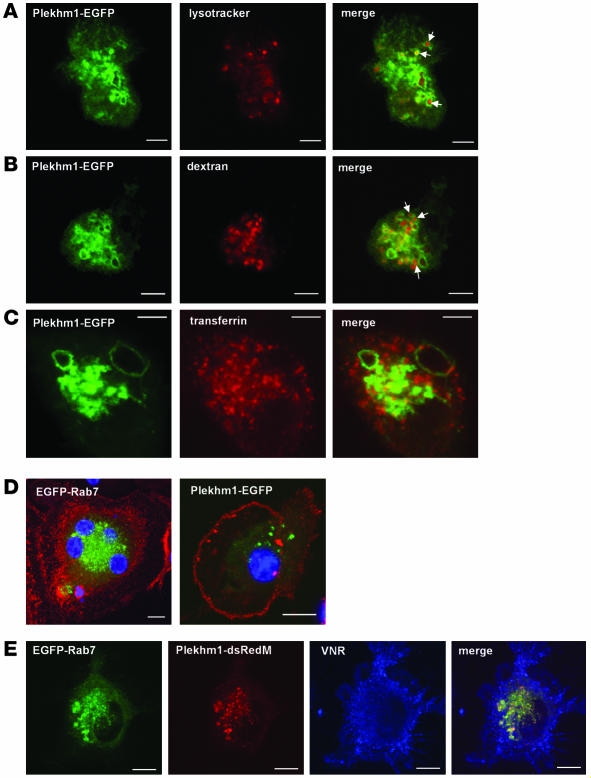
To confirm this localization of Plekhm1 in osteoclast-like cells, we transfected M-CSF–dependent macrophages derived from peripheral blood that had been stimulated to differentiate into osteoclasts by treatment with RANKL for 2 days. These cells were incubated in the presence of RANKL for a further 24 hours after transfection, resulting in cultures containing a proportion of VNR-positive mononuclear and multinucleated cells. Similar to the results in HEK cells, both EGFP-Rab7 and Plekhm1-EGFP localized to a vesicular compartment in VNR-positive cells (Figure 8D), which were often enlarged (Figure 8), and the 2 proteins colocalized to these vesicles in cotransfected cells (Figure 8E). The late endosomal/lysosomal nature of the Plekhm1-positive vesicles in these cells was confirmed using markers of the endocytic pathway, as most of the Plekhm1-expressing vesicles accumulated the acidotropic dye lysotracker, which accumulates in acidic vesicles of the endocytic pathway (Figure 8A), and endocytosed dextran, which had been pulse-chased and should therefore be in a late endosomal compartment (Figure 8B). By contrast, Plekhm1-positive vesicles did not accumulate endocytosed transferrin, a marker of recycling endosomes (Figure 8C). Interestingly, there was consistently less cytosolic Plekhm1 in prefusion osteoclasts than in HEK293 cells when the cells were transfected with Plekhm1-EGFP alone. Unfortunately, although continued culture with RANKL enabled the formation of resorbing osteoclasts expressing EGFP alone (i.e., empty vector), expression of Plekhm1-EGFP caused cell death within 48 hours of transfection, precluding determination of the localization of Plekhm1 in multinucleated, resorbing osteoclasts.
Figure 8. Plekhm1 colocalizes to late endosomes/lysosomes with Rab7 in prefusion osteoclasts.
(A–D) Prefusion human osteoclasts were transfected with Plekhm1-EGFP or EGFP-Rab7 and then analyzed 24 hours later by confocal microscopy. We also labeled acidic vesicles with lysotracker red (A), late endosomes/lysosomes with TRITC-dextran (B), and recycling endosomes with Alexa Fluor 633–transferrin (C) prior to fixation. Plekhm1 was associated mainly with intracellular vesicles that accumulated lysotracker and endocytosed dextran (A and B, arrows) but not transferrin, indicating that plekhm1 is localized to late endosomes/lysosomes. (D) Osteoclast-like cells were counterstained with an antibody to the VNR (red stain) and the nuclei stained with TO-PRO-3 iodide (blue stain). Both EGFP-Rab7 and Plekhm1-EGFP were localized to intracellular vesicles in VNR-positive osteoclasts. (E) Prefusion osteoclast cells were cotransfected with Plekhm1-dsRedM and EGFP-Rab7, then stained with an anti-VNR antibody. Plekhm1 and Rab7 almost completely colocalize on intracellular vesicles in VNR-positive prefusion osteoclast cells. Panels represent 1-μm xy optical sections. Scale bars: 5 μm.
Discussion
Bone resorption is an essential process for growing bone tissue, adaptating to exercise, and maintaining a healthy skeleton throughout life and is carried out by a highly specialized cell type, the osteoclast (53). Defects in the resorptive capacity of osteoclasts lead to osteopetrosis, an inherited disease manifested clinically by a generalized increase in bone mass and, in severe cases, accompanied by secondary bone marrow deficiency and neurologic problems due to cranial nerve compression (13). There is a wide range of clinical phenotypes in osteopetrosis, and it is becoming clear that these are associated with different genetic defects. It has long been recognized in studies of spontaneous animal models of osteopetrosis that the disease is heterogeneous, and it is now known that genetic mutations in any of the steps leading to osteoclast formation or activation of the resorption process can cause osteopetrosis (13). By contrast, the molecular mechanisms identified to play a role in human osteopetroses are so far completely restricted to alterations in osteoclast function (1). The 4 causative genes currently identified are all involved in the process of acidification of the extracellular compartment between the bone surface and the osteoclast. In these cases there is no deficiency in osteoclast formation, with normal or even increased numbers of osteoclasts present on the bone surface. In the case of recessive TCIRG1 and CLCN7 loss-of-function mutations, osteoclasts fail to polarize properly and, although rudimentary clear zones may be found, there is generally no sign of ruffled border formation, nor of extracellular matrix degradation (3, 5), indicating that loss of function of these genes completely abrogates the secretion of acid and proteolytic enzymes. In this report, we demonstrate for the first time to our knowledge that loss-of-function mutations in the PLEKHM1 gene underlie mild osteopetrosis in the ia rat as well as an intermediate form of human osteopetrosis. Moreover, we believe this to be the first indication of a function for Plekhm1 and highlights its crucial role in bone metabolism.
The expression of Plekhm1 protein is increased during RANKL-induced differentiation of osteoclasts and is much higher than in other cell types such as osteoblasts. Together with the fact that loss of function results in osteopetrosis, this indicates that the PLEKHM1 gene product is particularly important for osteoclast function, as is the case for other human osteopetrosis-causing genes. Microscopy analysis showed that osteoclasts from the osteopetrotic patient differentiated normally from PBMCs and attached to the surface of dentine by peripheral belts of podosomes. However, osteoclasts from the osteopetrotic patient — unlike those from her brother, who was heterozygous for the PLEKHM1 mutation — showed hardly any evidence of resorptive activity when cultured on dentine discs. These findings indicate that the Plekhm1 protein is not involved in the differentiation, attachment, or initial actin organization of osteoclasts, but rather in a later step of the bone resorption process. Although the formation of lysosomal vesicles was normal, the ruffled border was absent or underdeveloped in the patient-derived osteoclasts. The in vitro osteoclast phenotype we observed was very similar to that seen with other osteopetrotic mutations (54, 55). An interesting feature of osteoclasts from the patient with the PLEKHM1 mutation was an increased accumulation of the enzyme TRAP (which is highly expressed in osteoclasts), an effect also seen in the ia rat (15). Moreover, transmission EM of the osteoclasts from the 2 siblings homozygous for the PLEKHM1 mutation demonstrated the presence of electron-dense vesicles that were identical to those seen in the ia rat, which have previously been shown to contain TRAP (15). These vesicles were not seen in actively resorbing osteoclasts from the heterozygous brother, which instead contained many multivesicular bodies near the ruffled border. Such bodies were much less abundant in osteoclasts from the 2 homozygous siblings, suggesting that the multivesicular bodies contain material taken up from the resorption lacuna, whereas the dense granules contain TRAP and could be secretory vesicles. Unfortunately, we did not have a bone biopsy or sufficient patient-derived cultured osteoclasts to confirm expression of TRAP in these vesicles by immunoelectron microscopy, a technique that requires different fixation and embedding of tissue than the one used for routine transmission EM. Optimization of currently available anti-TRAP antibodies for such studies in human osteoclasts is underway. However, our findings suggest that the absence of Plekhm1 disrupts processes involved in the transport and/or fusion of late endosomal vesicles and consequently disrupts the formation of the ruffled border, which in turn impacts the endocytosis that normally occurs from this membrane domain (56). It is also possible that a defect in the maturation or trafficking of late endosomes prolongs their lifespan, allowing accumulation of their contents.
The exact function of Plekhm1 is unknown. Part of Plekhm1 has previously been described as the AP162 protein, which was suggested to function as an intracellular adaptor modulating apoptotic signals in colon tissue (23). However, the recombinant AP162 protein in the previously reported experiments lacks 130 amino acids at the N-terminal end of the Plekhm1 protein, indicating that full-length Plekhm1 has not been studied before. The putative role of the Plekhm1 protein in intracellular vesicular transport is supported by the presence of a RUN domain and 2 PH domains, which are found in proteins involved in intracellular signaling pathways of Ras-like small GTPases (29, 39, 40). In particular, the RUN domain has been identified in proteins that interact with small GTPases of the Rab family, which are master regulators of vesicular transport. These proteins include Iporin, a ubiquitously expressed protein that interacts with Rab1b (30), and Rabip4, a Rab4 effector that is associated with early endosomes (34). Rab GTPases are known to play a critical role in the regulation of osteoclast activity (57); in particular, Rab3D and Rab7 have been shown to be essential for osteoclastic resorption (58, 59).
We found further evidence for a role of the Plekhm1 protein in vesicular transport using overexpression studies in HEK293 cells, which demonstrated that Plekhm1 was partially associated with intracellular vesicles. These vesicles were identified as late endosomes or lysosomes, since Plekhm1 colocalizes with the late endosomal/lysosomal Rab7 and Rab9, but not with the early endosome–associated Rab5 or the Golgi-localized Rab6. Moreover, coexpression of Plekhm1 and Rab7 resulted in a striking redistribution of Plekhm1 to the Rab7-positive vesicles and a concomitant loss of the diffuse, cytoplasmic Plekhm1. This recruitment of Plekhm1 to late endosomes/lysosomes in cells expressing or overexpressing Rab7 is likely the result of an interaction between these proteins rather than changes in the nature of the compartment, since overexpression of another late endosomal/lysosomal Rab GTPase, Rab9, did not alter the distribution of Plekhm1, despite the fact that these proteins showed some colocalization. Importantly, we found that Plekhm1 also localized to late endosomes/lysosomes in prefusion osteoclasts, since it localized to acidic vesicles to which endocytosed dextran, but not transferrin (a marker of recycling endosomes), was transported. Moreover, Plekhm1 colocalized with Rab7 in VNR-positive prefusion osteoclasts, again suggesting an interaction between these 2 proteins. Interestingly, when transfected alone Plekhm1 showed a much more striking vesicular (and therefore less cytoplasmic) localization in these cells than in HEK293 cells, possibly because higher endogenous levels of Rab7 in prefusion osteoclasts enables greater association with late endosomes/lysosomes.
The correct localization of Rab GTPases to specific subcellular membranes is dependent on posttranslational prenylation, which involves the attachment of geranylgeranyl isoprenoid groups to 1 or, more commonly, 2 C-terminal cysteine residues by the enzyme Rab GGTase (50, 60, 61). Rab prenylation can be blocked selectively using the specific Rab GGTase inhibitor 3-PEHPC, thereby preventing the membrane localization of Rab proteins (62). Inhibition of Rab prenylation with 3-PEHPC disrupted the vesicular localization of both Rab7 and Plekhm1 without affecting the distribution of acidic vesicles, indicating that Plekhm1 requires Rab prenylation for its late endosomal/lysosomal localization and therefore most likely interacts with Rab7 on these vesicles. Finally, we studied the distribution of Plekhm1 when coexpressed with GTPase-deficient (active) or dominant-negative mutants of Rab7, which are predominantly associated with vesicular structures (similar to wild-type Rab7) or localized in the cytosol, respectively (41). Whereas Plekhm1 colocalized with GTPase-deficient Rab7 on intracellular vesicles, coexpression of the dominant-negative Rab7 mutant and Plekhm1 resulted in loss of the endosomal localization of both Rab7 and Plekhm1, offering further evidence that Plekhm1 interacts with Rab7 and is recruited to late endosomes/lysosomes by prenylated, GTP-bound Rab7.
Recruitment to late endosomes/lysosomes is a characteristic that Plekhm1 shares with other proteins that have been identified as Rab7 effectors, such as Rab-interacting lysosomal protein (RILP) and Rabring7 (63, 64). However, these Rab7-interacting proteins also cause perinuclear clustering of the Rab7-positive vesicles (implicating them in transport of these vesicles), an effect that we did not see in Plekhm1-overexpressing cells. Since Plekhm1 overexpression caused enlargement of these endosomes, it is possible that it is involved in Rab7-mediated fusion of these vesicles rather than their transport.
It has previously been shown that Rab7 is necessary for osteoclast function, since blocking Rab7 expression in osteoclasts using antisense oligodeoxynucleotides disrupted actin ring formation and targeting of vesicles to the ruffled border and furthermore inhibited bone resorption in vitro (59). These effects may be mediated by altered activity of Rac1, which has recently been suggested to be a direct effector of Rab7 in osteoclasts (65). Interestingly, the defective ruffled border and extended clear zone that is seen in osteoclasts lacking Rab7 (59) are also characteristic features of osteoclasts in the ia rat (15), offering further evidence that Rab7 and Plekhm1 are involved in the same vesicular transport processes in osteoclasts. However, although the osteoclasts generated from the patient with the PLEKHM1 mutation clearly had a defective ruffled border, we could not discern any extension of the clear zone.
Although the exact role of Plekhm1 in bone resorption will require further investigation, our data strongly implicate Plekhm1 as a component of Rab7-regulated late endosomal trafficking in osteoclasts. Determining the exact function of the Plekhm1 protein may reveal new therapeutic approaches to osteopetrosis, osteoporosis, and other osteoclast diseases. Although there was no evidence of clinical symptoms other than osteopetrosis in the patient lacking Plekhm1 protein, it remains to be determined whether Plekhm1 plays a role in vesicular transport in other cell types.
Methods
Animals.
Mutant ia rats were obtained from inbred colonies maintained at the University of Massachusetts Medical School under specific pathogen–free conditions. Radiography within 3 days of birth was used to identify homozygous mutant ia animals by the failure to develop marrow cavities in the long bones. All procedures were in accordance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of the University of Massachusetts Medical School. Outbreeding of animals of the ia stock was performed as described previously (20).
Analysis of genetic markers.
Genomic DNA was isolated from tail biopsies of F2, F4, F6, or F8 mutants and used to refine the ia candidate region. Markers were selected from the Ensembl rat genome browser (http://www.ensembl.org/Rattus_norvegicus/) and analyzed by PCR using fluorescently labeled primers, and the fragments were separated on an ABI 3100 genetic analyzer (Applied Biosystems).
RNA isolation and RT-PCR.
Total RNA was isolated using TRI zol reagent (Invitrogen). cDNA synthesis was performed, starting from 5 μg total RNA primed with oligo(dT) using the SuperScript III first-strand synthesis system (Invitrogen).
Sequence analysis.
The Big Dye Terminator v1.1 Cycle Sequencing Kit was used, and the PCR fragments were analyzed on an ABI 3100 genetic analyzer (both from Applied Biosystems).
Expression analysis.
Expression of the rat Plekhm1 gene and the existence of alternative splice variants were analyzed with a rat Multiple Tissue cDNA panel (Clontech). PCR primers were developed to amplify the complete coding sequence of the rat Plekhm1 gene. Detection of the Plekhm1 protein was carried out by Western blotting. Anti-Plekhm1 antibodies were raised in rabbits against 2 peptides (PTSPKGKSWISEDDFC, amino acids 431–445; and LQKVRPQQEEEWVNIC, amino acids 627–640) of the rat Plekhm1 protein using a standard immunization protocol (Eurogentec). Lysates were prepared from the following: (a) human osteoblasts isolated and cultured from the femoral head of osteoporotic patients receiving a hip replacement; (b) human osteoclasts generated from peripheral blood of healthy volunteers treated for different time periods with RANKL; and (c) osteoclasts generated from the osteopetrotic patient. As a positive control, HEK293 cells were transfected with the Plekhm1-FLAG construct and incubated for 24 hours prior to lysis. Cell lysates (50 μg) were separated by electrophoresis on 10% criterion gels (Bio-Rad), transferred to PVDF membrane, and then blotted using 1:1,000 dilution of the Plekhm1 antibody. Bands were visualized as described previously (51), and densitometry was performed from 4 separate Western blots using Quantity-One software (version 4.5.1; Bio-Rad). Since cross-reactivity with other proteins was found on Western blot analysis, this antibody was not used for immunohistochemistry or immunofluorescence analysis. This study was approved by the ethical committee of the University of Antwerp. Subjects gave informed consent for the study.
Generation of osteoclasts from human PBMCs.
Human osteoclasts were generated as previously described (66). After a total of 15 days’ incubation, cells were fixed in either 4% paraformaldehyde in 0.1 M phosphate buffer (for light microscopy) or in glutaraldehyde (for EM). In some cases, acidic vesicles were stained by incubation with the fixable acidotropic probe Lysotracker Red DND-99 (0.5 μM; Invitrogen) in fresh medium for 30 minutes prior to fixation.
Immunostaining.
The fixed osteoclasts were permeabilized with 0.2% Triton X-100 then incubated with 10% FCS in PBS to block nonspecific binding. Cells were then incubated with primary antibody against the VNR (23C6; courtesy of M. Horton, University College London, London, United Kingdom) for 1 hour at room temperature. After washing 4 times with PBS, the cells were incubated with Alexa Fluor 633–conjugated anti-mouse IgG (Invitrogen) in 5% FCS/PBS for 1 hour at room temperature. F-actin rings were visualized by staining with 0.5 μg/ml tetramethylrhodamine isothiocyanate–labeled (TRITC-labeled) or FITC-labeled phalloidin (Sigma-Aldrich), and the nuclei were stained with 0.5 μg/ml Sytox Green (Invitrogen). The dentine surface was visualized by labeling with a fluorescent bisphosphonate derivative, FL-ALN (20 μM) (67). Cells were analyzed by confocal microscopy using a Zeiss LSM 510 confocal microscope equipped with argon (excitation, 488 nm) and helium-neon (excitation, 543 and 633 nm) lasers. Osteoclasts were also stained for TRAP using naphthol-ASBI phosphate substrate as described previously (68) and analyzed by light microscopy using a Zeiss Axiovert microscope equipped with an Optronics charge-coupled device camera (model DEI-750).
EM.
Transmission EM was performed as previously described (51). For scanning EM, osteoclast cultures were fixed in 2% glutaraldehyde in 0.1 M sodium cacodylate plus 0.1 M sucrose for 24 hours at 4°C. Specimens were washed twice in the same buffer and then submerged in a filtered solution of 1% tannic acid in 0.15 M cacodylate for 1 hour. After 3 short washes in 0.1 M buffer, the samples were postfixed in 0.5% osmium tetroxide in 0.1 M cacodylate buffer, and after another 3 short washes in buffer, they were dehydrated through graded ethanols, finishing with 3 changes of 100% ethanol. Slices were then placed in hexamethyldisilasane for 3 minutes before being moved to a desiccator for 25 minutes to avoid water contamination. Specimens were coated with gold before being examined in a Jeol 35S scanning electron microscope at 10 kV. Images were taken using SemAfore software (version 5.01; Jeol). This preparation procedure provided better results (especially less shrinkage) than routine fixation and dehydration with critical point drying.
DNA constructs.
Human PLEKHM1 cDNA was amplified by PCR, starting from the IMAGE cDNA clone 5535600 (RZPD) and subcloned in the pEGFP-N1, pdsRedMonomer-N1 vector (Clontech) and FLAG-pcDNA3 vector. The mutant rat and human Plekhm1-dsRedM constructs consisted of 1,026 and 297 nucleotides, respectively. The EGFP-Rab5, -Rab6, and -Rab7 constructs were kindly provided by M. Seabra (Imperial College London, London, United Kingdom). Rab9-EGFP was provided by M. Fukuda (RIKEN, Saitama, Japan). The EGFP–Rab7-Q67L and –Rab7-T22N constructs were kindly provided by B. van Deurs (University of Copenhagen, Copenhagen, Denmark).
Transfection of HEK293 cells.
HEK293 cells were grown in DMEM supplemented with 10% FCS and seeded onto 13-mm glass coverslips in 24-well plates at 1 × 105 cells/well. Cells were transfected with 200 ng of purified plasmid using Fugene 6 transfection reagent (Roche). In the case of dual transfections, 100 ng of each plasmid was used. In some cases, cells were also incubated with 1 mM 3-PEHPC, a specific Rab GGTase inhibitor (51, 62). Cells were incubated for 24 hours, and in some cases acidic vesicles were labeled using lysotracker as described above, prior to fixation in 4% formaldehyde. Cells were examined by confocal microscopy after mounting the coverslips onto glass slides.
Transfection of prefusion osteoclasts.
M-CSF–dependent macrophages were generated from healthy volunteers and transfected as previously described (69). Briefly, cultures of M-CSF–dependent macrophages were stimulated with 100 ng/ml recombinant human RANKL (rhRANKL; PeproTech) for 2 days prior to transfection. Cells were then detached by scraping after incubation in 1 mg/ml trypsin for 15 minutes, and 1 × 106 cells were electroporated with 2 μg endotoxin-free DNA (EGFP-Rab7, Plekhm1-EGFP, or Plekhm1-dsRedM) in Macrophage Nucleofector Solution (Amaxa Biosystems) using an Amaxa Nucleofector II according to the manufacturer’s instructions. Cells were diluted in α-MEM (supplemented with 20% FCS, 100 U/ml penicillin, 0.1 mg/ml streptomycin, 2 mM l-glutamine, 20 ng/ml rhMCSF, and 100 ng/ml rhRANKL) and seeded onto 9-mm glass coverslips in 48-well plates (1 × 105 cells/well). Twenty-four hours later, cells were fixed in 4% formaldehyde. In some cases, acidic vesicles were labeled using lysotracker as described above, and recycling endosomes were labeled by incubating cells with 20 μg/ml Alexa Fluor 633–transferrin conjugate for 30 minutes in fresh medium prior to fixation. Labeling of late endosomes/lysosomes was carried out by pulse-chase studies with dextran, in which cells were incubated for 30 minutes with 0.25 mg/ml lysine-fixable TRITC-dextran (10 kDa), then washed and incubated in fresh medium for a further 30 minutes prior to fixation. Osteoclast-like cells were counterstained by incubation with the specific anti-VNR antibody 23C6 described above in PBS with 5% FCS for 1 hour, followed by incubation with 1:200 anti-mouse IgG (conjugated to Alexa Fluor 633 or Alexa Fluor 594) for 1 hour. Nuclei were visualized by staining with 0.5 μM TO-PRO-3 iodide (Invitrogen) for 10 minutes. Coverslips were mounted onto glass slides and analyzed by confocal microscopy as described above.
Acknowledgments
The authors thank Jan Van Daele for technical assistance with the confocal imaging, Martine Biervliet for taking blood samples, and Kevin Mackenzie, John Greenhorn, Adam Taylor, and Denise Tosh for technical assistance with osteoclast cultures and EM. L. Van Wesenbeeck is a postdoctoral researcher of the Fonds voor Wetenschappelijk Onderzoek (FWO). This work was supported by an exchange scholarship grant from the European Calcified Tissue Society and FWO grant 1.5.032.07N (to L. Van Wesenbeeck); by FWO grant G0117.06, FP5 EU project GENOMOS (QLK6-CT-2002-02629), and BOF-NOI project from the University of Antwerp (to W. Van Hul); by National Institute of Dental and Craniofacial Research NIH grant DEO7444 (to P.R. Odgren); by “Fondo per gli Investimenti per la Ricerca di Base” (FIRB) grant RBAU01X3NH and Telethon grant GGP06019 (to A. Teti); by FIRB grants RBNE019J9W-004 and RBIN04CHXT and Fondazione Cariplo, Progetto N.o.b.e.l. (to A. Villa); by a career establishment award from the European Calcified Tissue Society (to M.H. Helfrich); and by Research Councils UK (to F.P. Coxon).
Footnotes
Nonstandard abbreviations used: EGFP, enhanced GFP; GGTase, geranylgeranyl transferase; ia, incisors absent; PH, pleckstrin homology; PLEKHM1, pleckstrin homology domain–containing family M (with RUN domain) member 1; TRAP, tartrate-resistant acid phosphatase; TRITC, tetramethylrhodamine isothiocyanate; VNR, vitronectin receptor.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:919–930 (2007). doi:10.1172/JCI30328
Liesbeth Van Wesenbeeck, Paul R. Odgren, and Fraser P. Coxon contributed equally to this work.
References
- 1.Balemans W., Van Wesenbeeck L., Van Hul W. A clinical and molecular overview of the human osteopetroses. Calcif. Tissue Int. 2005;77:263–274. doi: 10.1007/s00223-005-0027-6. [DOI] [PubMed] [Google Scholar]
- 2.Sly W.S., Hewett-Emmett D., Whyte M.P., Yu Y.S., Tashian R.E. Carbonic anhydrase II deficiency identified as the primary defect in the autosomal recessive syndrome of osteopetrosis with renal tubular acidosis and cerebral calcification. Proc. Natl. Acad. Sci. U. S. A. 1983;80:2752–2756. doi: 10.1073/pnas.80.9.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kornak U., et al. Mutations in the a3 subunit of the vacuolar H(+)-ATPase cause infantile malignant osteopetrosis. Hum. Mol. Genet. 2000;9:2059–2063. doi: 10.1093/hmg/9.13.2059. [DOI] [PubMed] [Google Scholar]
- 4.Frattini A., et al. Defects in TCIRG1 subunit of the vacuolar proton pump are responsible for a subset of human autosomal recessive osteopetrosis. Nat. Genet. 2000;25:343–346. doi: 10.1038/77131. [DOI] [PubMed] [Google Scholar]
- 5.Kornak U., et al. Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell. 2001;104:205–215. doi: 10.1016/s0092-8674(01)00206-9. [DOI] [PubMed] [Google Scholar]
- 6.Cleiren E., et al. Albers-Schonberg disease (autosomal dominant osteopetrosis, type II) results from mutations in the ClCN7 chloride channel gene. Hum. Mol. Genet. 2001;10:2861–2867. doi: 10.1093/hmg/10.25.2861. [DOI] [PubMed] [Google Scholar]
- 7.Campos-Xavier A.B., Saraiva J.M., Ribeiro L.M., Munnich A., Cormier-Daire V. Chloride channel 7 (CLCN7) gene mutations in intermediate autosomal recessive osteopetrosis. Hum. Genet. 2003;112:186–189. doi: 10.1007/s00439-002-0861-9. [DOI] [PubMed] [Google Scholar]
- 8.Chalhoub N., et al. Grey-lethal mutation induces severe malignant autosomal recessive osteopetrosis in mouse and human. Nat. Med. 2003;9:399–406. doi: 10.1038/nm842. [DOI] [PubMed] [Google Scholar]
- 9.Ramirez A., et al. Identification of a novel mutation in the coding region of the grey-lethal gene OSTM1 in human malignant infantile osteopetrosis. Hum. Mutat. 2004;23:471–476. doi: 10.1002/humu.20028. [DOI] [PubMed] [Google Scholar]
- 10.Quarello P., et al. Severe malignant osteopetrosis caused by a gl gene mutation. J. Bone Miner. Res. 2004;19:1194–1199. doi: 10.1359/JBMR.040407. [DOI] [PubMed] [Google Scholar]
- 11.Pangrazio A., et al. Mutations in OSTM1 (grey lethal) define a particularly severe form of autosomal recessive osteopetrosis with neural involvement. J. Bone Miner. Res. 2006;21:1098–1105. doi: 10.1359/jbmr.060403. [DOI] [PubMed] [Google Scholar]
- 12.Lange P.F., Wartosch L., Jentsch T.J., Fuhrmann J.C. ClC-7 requires Ostm1 as a beta-subunit to support bone resorption and lysosomal function. Nature. 2006;440:220–223. doi: 10.1038/nature04535. [DOI] [PubMed] [Google Scholar]
- 13.Van Wesenbeeck L., Van Hul W. Lessons from osteopetrotic mutations in animals: impact on our current understanding of osteoclast biology. Crit. Rev. Eukaryot. Gene Expr. 2005;15:133–162. doi: 10.1615/critreveukaryotgeneexpr.v15.i2.40. [DOI] [PubMed] [Google Scholar]
- 14.Greep R.O. An hereditary absence of the incisors teeth. J. Hered. 1941;32:397–398. [Google Scholar]
- 15.Reinholt F.P., et al. Extensive clear zone and defective ruffled border formation in osteoclasts of osteopetrotic (ia/ia) rats: implications for secretory function. Exp. Cell Res. 1999;251:477–491. doi: 10.1006/excr.1999.4585. [DOI] [PubMed] [Google Scholar]
- 16.Marks S.C., Jr. Pathogenesis of osteopetrosis in the ia rat: reduced bone resorption due to reduced osteoclast function. Am. J. Anat. 1973;138:165–189. doi: 10.1002/aja.1001380204. [DOI] [PubMed] [Google Scholar]
- 17.Marks S.C., Jr. Osteopetrosis in the ia rat cured by spleen cells from a normal littermate. Am. J. Anat. 1976;146:331–338. doi: 10.1002/aja.1001460308. [DOI] [PubMed] [Google Scholar]
- 18.Miller S.C., Marks S.C., Jr. Osteoclast kinetics in osteopetrotic (ia) rats cured by spleen cell transfers from normal littermates. Calcif. Tissue Int. 1982;34:422–427. doi: 10.1007/BF02411278. [DOI] [PubMed] [Google Scholar]
- 19.Marks S.C., Jr. Studies of the mechanism of spleen cell cure for osteopetrosis in ia rats: appearance of osteoclasts with ruffled borders. Am. J. Anat. 1978;151:119–129. doi: 10.1002/aja.1001510110. [DOI] [PubMed] [Google Scholar]
- 20.Van Wesenbeeck L., Odgren P.R., Mackay C.A., Van Hul W. Localization of the gene causing the osteopetrotic phenotype in the incisors absent (ia) rat on chromosome 10q32.1. J. Bone Miner. Res. 2004;19:183–189. doi: 10.1359/jbmr.2004.19.2.183. [DOI] [PubMed] [Google Scholar]
- 21.Raber S.A. The dense metaphyseal band sign. Radiology. 1999;211:773–774. doi: 10.1148/radiology.211.3.r99jn13773. [DOI] [PubMed] [Google Scholar]
- 22.Jurdic P., Saltel F., Chabadel A., Destaing O. Podosome and sealing zone: specificity of the osteoclast model. Eur. J. Cell Biol. 2006;85:195–202. doi: 10.1016/j.ejcb.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Hartel-Schenk S., et al. Novel adapter protein AP162 connects a sialyl-Le(x)-positive mucin with an apoptotic signal transduction pathway. Glycoconj. J. 2001;18:915–923. doi: 10.1023/a:1022256610674. [DOI] [PubMed] [Google Scholar]
- 24.Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 25.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 26.Mulder N.J., et al. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 2003;31:315–318. doi: 10.1093/nar/gkg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorick K.L., et al. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ono Y., et al. Phorbol ester binding to protein kinase C requires a cysteine-rich zinc-finger-like sequence. Proc. Natl. Acad. Sci. U. S. A. 1989;86:4868–4871. doi: 10.1073/pnas.86.13.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Callebaut I., de Gunzburg J., Goud B., Mornon J.P. RUN domains: a new family of domains involved in Ras-like GTPase signaling. Trends Biochem. Sci. 2001;26:79–83. doi: 10.1016/s0968-0004(00)01730-8. [DOI] [PubMed] [Google Scholar]
- 30.Bayer M., et al. Identification and characterization of Iporin as a novel interaction partner for rab1. BMC Cell Biol. 2005;6:15. doi: 10.1186/1471-2121-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janoueix-Lerosey I., Jollivet F., Camonis J., Marche P.N., Goud B. Two-hybrid system screen with the small GTP-binding protein Rab6. Identification of a novel mouse GDP dissociation inhibitor isoform and two other potential partners of Rab6. J. Biol. Chem. 1995;270:14801–14808. doi: 10.1074/jbc.270.24.14801. [DOI] [PubMed] [Google Scholar]
- 32.Janoueix-Lerosey I., Pasheva E., de Tand M.F., Tavitian A., de Gunzburg J. Identification of a specific effector of the small GTP-binding protein Rap2. Eur. J. Biochem. 1998;252:290–298. doi: 10.1046/j.1432-1327.1998.2520290.x. [DOI] [PubMed] [Google Scholar]
- 33.Yang J., Kim O., Wu J., Qiu Y. Interaction between tyrosine kinase Etk and a RUN domain- and FYVE domain-containing protein RUFY1. A possible role of ETK in regulation of vesicle trafficking. J. Biol. Chem. 2002;277:30219–30226. doi: 10.1074/jbc.M111933200. [DOI] [PubMed] [Google Scholar]
- 34.Mari M., Macia E., Le Marchand-Brustel Y., Cormont M. Role of the FYVE finger and the RUN domain for the subcellular localization of Rabip4. J. Biol. Chem. 2001;276:42501–42508. doi: 10.1074/jbc.M104885200. [DOI] [PubMed] [Google Scholar]
- 35.Cozier G.E., Carlton J., Bouyoucef D., Cullen P.J. Membrane targeting by pleckstrin homology domains. Curr. Top. Microbiol. Immunol. 2004;282:49–88. doi: 10.1007/978-3-642-18805-3_3. [DOI] [PubMed] [Google Scholar]
- 36.Lemmon M.A. Pleckstrin homology domains: not just for phosphoinositides. Biochem. Soc. Trans. 2004;32:707–711. doi: 10.1042/BST0320707. [DOI] [PubMed] [Google Scholar]
- 37.Maghazachi A.A. Intracellular signaling events at the leading edge of migrating cells. Int. J. Biochem. Cell Biol. 2000;32:931–943. doi: 10.1016/s1357-2725(00)00035-2. [DOI] [PubMed] [Google Scholar]
- 38.Lemmon M.A., Ferguson K.M., Abrams C.S. Pleckstrin homology domains and the cytoskeleton. FEBS Lett. 2002;513:71–76. doi: 10.1016/s0014-5793(01)03243-4. [DOI] [PubMed] [Google Scholar]
- 39.Ingley E., Hemmings B.A. Pleckstrin homology (PH) domains in signal transduction. J. Cell. Biochem. 1994;56:436–443. doi: 10.1002/jcb.240560403. [DOI] [PubMed] [Google Scholar]
- 40.Johansson M., Lehto M., Tanhuanpaa K., Cover T.L., Olkkonen V.M. The oxysterol-binding protein homologue ORP1L interacts with Rab7 and alters functional properties of late endocytic compartments. Mol. Biol. Cell. 2005;16:5480–5492. doi: 10.1091/mbc.E05-03-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bucci C., Thomsen P., Nicoziani P., McCarthy J., van Deurs B. Rab7: a key to lysosome biogenesis. Mol. Biol. Cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mesa R., Salomon C., Roggero M., Stahl P.D., Mayorga L.S. Rab22a affects the morphology and function of the endocytic pathway. J. Cell Sci. 2001;114:4041–4049. doi: 10.1242/jcs.114.22.4041. [DOI] [PubMed] [Google Scholar]
- 43.Barbieri M.A., Li G., Mayorga L.S., Stahl P.D. Characterization of Rab5:Q79L-stimulated endosome fusion. Arch. Biochem. Biophys. 1996;326:64–72. doi: 10.1006/abbi.1996.0047. [DOI] [PubMed] [Google Scholar]
- 44.Bucci C., et al. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- 45.Goud B., Zahraoui A., Tavitian A., Saraste J. Small GTP-binding protein associated with Golgi cisternae. Nature. 1990;345:553–556. doi: 10.1038/345553a0. [DOI] [PubMed] [Google Scholar]
- 46.Vitelli R., et al. Role of the small GTPase Rab7 in the late endocytic pathway. J. Biol. Chem. 1997;272:4391–4397. doi: 10.1074/jbc.272.7.4391. [DOI] [PubMed] [Google Scholar]
- 47.Meresse S., Gorvel J.P., Chavrier P. The rab7 GTPase resides on a vesicular compartment connected to lysosomes. J. Cell Sci. 1995;108:3349–3358. doi: 10.1242/jcs.108.11.3349. [DOI] [PubMed] [Google Scholar]
- 48.Feng Y., Press B., Wandinger-Ness A. Rab 7: an important regulator of late endocytic membrane traffic. J. Cell Biol. 1995;131:1435–1452. doi: 10.1083/jcb.131.6.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lombardi D., et al. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 1993;12:677–682. doi: 10.1002/j.1460-2075.1993.tb05701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casey P.J., Seabra M.C. Protein prenyltransferases. J. Biol. Chem. 1996;271:5289–5292. doi: 10.1074/jbc.271.10.5289. [DOI] [PubMed] [Google Scholar]
- 51.Coxon F.P., et al. Identification of a novel phosphonocarboxylate inhibitor of Rab geranylgeranyl transferase that specifically prevents Rab prenylation in osteoclasts and macrophages. J. Biol. Chem. 2001;276:48213–48222. doi: 10.1074/jbc.M106473200. [DOI] [PubMed] [Google Scholar]
- 52.Ceresa B.P., Bahr S.J. rab7 activity affects epidermal growth factor:epidermal growth factor receptor degradation by regulating endocytic trafficking from the late endosome. J. Biol. Chem. 2006;281:1099–1106. doi: 10.1074/jbc.M504175200. [DOI] [PubMed] [Google Scholar]
- 53.Vaananen K. Mechanism of osteoclast mediated bone resorption--rationale for the design of new therapeutics. Adv. Drug Deliv. Rev. 2005;57:959–971. doi: 10.1016/j.addr.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 54.Del Fattore A., et al. Clinical, genetic, and cellular analysis of 49 osteopetrotic patients: implications for diagnosis and treatment. J. Med. Genet. 2006;43:315–325. doi: 10.1136/jmg.2005.036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taranta A., et al. Genotype-phenotype relationship in human ATP6i-dependent autosomal recessive osteopetrosis. Am. J. Pathol. 2003;162:57–68. doi: 10.1016/S0002-9440(10)63798-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Palokangas H., Mulari M., Vaananen H.K. Endocytic pathway from the basal plasma membrane to the ruffled border membrane in bone-resorbing osteoclasts. J. Cell Sci. 1997;110:1767–1780. doi: 10.1242/jcs.110.15.1767. [DOI] [PubMed] [Google Scholar]
- 57.Coxon F.P., Rogers M.J. The role of prenylated small GTP-binding proteins in the regulation of osteoclast function. Calcif. Tissue Int. 2003;72:80–84. doi: 10.1007/s00223-002-2017-2. [DOI] [PubMed] [Google Scholar]
- 58.Pavlos N.J., et al. Rab3D regulates a novel vesicular trafficking pathway that is required for osteoclastic bone resorption. Mol. Cell. Biol. 2005;25:5253–5269. doi: 10.1128/MCB.25.12.5253-5269.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao H., Laitala-Leinonen T., Parikka V., Vaananen H.K. Downregulation of small GTPase Rab7 impairs osteoclast polarization and bone resorption. J. Biol. Chem. 2001;276:39295–39302. doi: 10.1074/jbc.M010999200. [DOI] [PubMed] [Google Scholar]
- 60.Pfeffer S. A model for Rab GTPase localization. Biochem. Soc. Trans. 2005;33:627–630. doi: 10.1042/BST0330627. [DOI] [PubMed] [Google Scholar]
- 61.Pfeffer S., Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat. Rev. Mol. Cell Biol. 2004;5:886–896. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- 62.Coxon F.P., et al. Phosphonocarboxylate inhibitors of Rab geranylgeranyl transferase disrupt the prenylation and membrane localization of Rab proteins in osteoclasts in vitro and in vivo. Bone. 2005;37:349–358. doi: 10.1016/j.bone.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 63.Cantalupo G., Alifano P., Roberti V., Bruni C.B., Bucci C. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 2001;20:683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mizuno K., Kitamura A., Sasaki T. Rabring7, a novel Rab7 target protein with a RING finger motif. Mol. Biol. Cell. 2003;14:3741–3752. doi: 10.1091/mbc.E02-08-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun Y., Buki K.G., Ettala O., Vaaraniemi J.P., Vaananen H.K. Possible role of direct Rac1-Rab7 interaction in ruffled border formation of osteoclasts. J. Biol. Chem. 2005;280:32356–32361. doi: 10.1074/jbc.M414213200. [DOI] [PubMed] [Google Scholar]
- 66.Dunford J.E., Rogers M.J., Ebetino F.H., Phipps R.J., Coxon F.P. Inhibition of protein prenylation by bisphosphonates causes sustained activation of Rac, Cdc42, and Rho GTPases. J. Bone Miner. Res. 2006;21:684–694. doi: 10.1359/jbmr.060118. [DOI] [PubMed] [Google Scholar]
- 67.Thompson K., Rogers M.J., Coxon F.P., Crockett J.C. Cytosolic entry of bisphosphonate drugs requires acidification of vesicles after fluid-phase endocytosis. Mol. Pharmacol. 2006;69:1624–1632. doi: 10.1124/mol.105.020776. [DOI] [PubMed] [Google Scholar]
- 68.van’t Hof R.J., Tuinenburg-Bol Raap A.C., Nijweide P.J. Induction of osteoclast characteristics in cultured avian blood monocytes; modulation by osteoblasts and 1,25-(OH)2 vitamin D3. Int. J. Exp. Pathol. 1995;76:205–214. [PMC free article] [PubMed] [Google Scholar]
- 69.Taylor A., Rogers M.J., Tosh D., Coxon F.P. A novel method for efficient generation of transfected human osteoclasts. Calcif. Tissue Int. 2007;80:132–136. doi: 10.1007/s00223-006-0245-6. [DOI] [PubMed] [Google Scholar]