Abstract
The Saccharomyces cerevisiae Cdc6 protein is crucial for DNA replication. In the absence of cyclin-dependent kinase (CDK) activity, Cdc6 binds to replication origins, and loads Mcm proteins. In the presence of CDK activity, Cdc6 does not bind to origins, and this helps prevent rereplication. CDK activity affects Cdc6 function by multiple mechanisms: CDK activity affects transcription of CDC6, degradation of Cdc6, nuclear import of Cdc6, and binding of Cdc6 to Clb2. Here we examine some of these mechanisms individually. We find that when Cdc6 is forced into the nucleus during late G1 or S, it will not substantially reload onto chromatin no matter whether its CDK sites are present or not. In contrast, at a G2/M nocodazole arrest, Cdc6 will reload onto chromatin if and only if its CDK sites have been removed. Trace amounts of nonphosphorylatable Cdc6 are dominant lethal in strains bearing nonphosphorylatable Orc2 and Orc6, apparently because of rereplication. This synthetic dominant lethality occurs even in strains with wild-type MCM genes. Nonphosphorylatable Cdc6, or Orc2 and Orc6, sensitize cells to rereplication caused by overexpression of various replication initiation proteins such as Dpb11 and Sld2.
INTRODUCTION
Assembly of prereplication complexes (pre-RCs) at future origins is necessary for initiation of DNA replication (Diffley, 1996; Stillman, 1996; Kelly and Brown, 2000; Bell and Dutta, 2002). In the yeast Saccharomyces cerevisiae, but also in many other organisms, the binding of Cdc6 to the origin recognition complex (ORC) is a critical step in the formation of pre-RCs (Liang et al., 1995; Cocker et al., 1996; Detweiler and Li, 1997) and essential for subsequent loading of Mcm proteins (Mcm2-7; Piatti et al., 1996; Santocanale and Diffley, 1996; Aparicio et al., 1997; Donovan et al., 1997; Tanaka et al., 1997). After licensing the origin by loading Mcms, endogenous Cdc6 dissociates from the replicative complex and only reassociates with chromatin late in M-phase (Piatti et al., 1996; Weinreich et al., 1999) when cyclin-dependent protein kinase (Clb-Cdc28) activities are absent. Although it is clear that loading of Cdc6 onto chromatin is inhibited by cyclin-dependent kinase (CDK) activity, the mechanism of inhibition is complex, and there are many modes of regulation (see Figure 1). Expression of CDC6 is cell cycle regulated (Zwerschke et al., 1994; Piatti et al., 1995) and therefore is at least indirectly controlled by CDK activity, such that CDC6 is expressed in late M and in G1, but is not expressed in S, G2, and early M. Phosphorylation of Cdc6 promotes ubiquitin-mediated proteolysis of Cdc6 (Drury et al., 1997; Elsasser et al., 1999; Sanchez et al., 1999; Drury et al., 2000). Phosphorylation of Cdc6 near its N-terminal nuclear localization signal may inhibit nuclear import (Jong et al., 1996). These three modes of regulation—transcription, proteolysis, and nuclear exclusion—work to minimize the amount of Cdc6 in the nucleus of a G2/early M-phase cell. In addition, in vitro experiments of Mimura et al. (2004) suggest that even if Cdc6 were present in the nucleus of an M-phase cell, it might not bind to chromatin. In these experiments, Mimura et al. found that wild-type (i.e., phosphorylatable) Cdc6 could not bind to beads containing ARS1 DNA in G2/M extracts, whereas mutant, nonphosphorylatable Cdc6 could bind (Mimura et al., 2004). In contrast to these in vitro results, Tanaka et al. (1997) found that in vivo, ectopically expressed Cdc6 could bind to chromatin in G2/M; however in these experiments CDC6 was overexpressed from the GAL promoter.
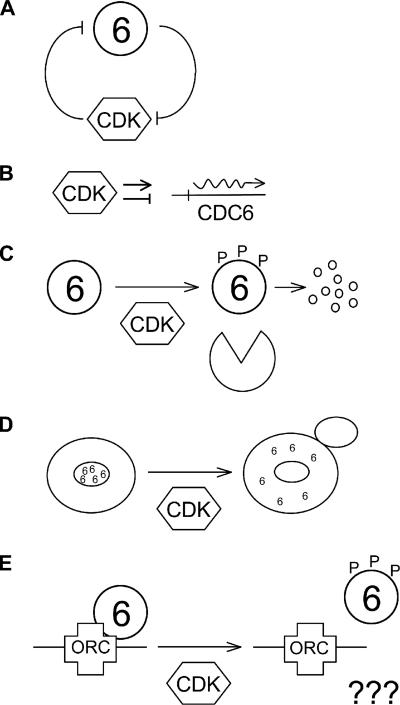
Figure 1.
CDK activity controls Cdc6 and vice versa. (A) CDK activity down-regulates Cdc6, but Cdc6 inhibits CDK activity. Thus CDK and Cdc6 form a negative feedback loop. (B) CDK activity both activates and represses CDC6 transcription, depending on the time of the cell cycle. (C) Phosphorylation of Cdc6 by CDK causes proteolysis of Cdc6. (D) Phosphorylation of Cdc6 by CDK may cause nuclear exclusion of Cdc6. (E) Phosphorylation of Cdc6 by CDK may directly interfere with binding of Cdc6 to ORC.
These various mechanisms contribute to regulation of Cdc6 function by CDK. It has been difficult to assess the relative importance of different mechanisms of regulation for at least two kinds of reasons. First, the fact that multiple mechanisms of regulation are interleaved and dependent on CDK activity. Second, Cdc6 is itself a CDK inhibitor (Bueno and Russell, 1992; Elsasser et al., 1996; Calzada et al., 2001), so any manipulation that results in relatively high Cdc6 abundance has the ability to inhibit CDK activity and feedback on multiple mechanisms of regulation (Figure 1). These difficulties are exemplified in the studies of Tanaka et al. (1997), who found that ectopically expressed Cdc6 could reload onto chromatin in G2/M. At face value, this result suggests that the normal failure of Cdc6 to reload is controlled mainly at the level of expression. However, the Cdc6 in these studies was overexpressed from the GAL promoter and could have inhibited CDK activity, thus indirectly affecting multiple modes of regulation. Furthermore, Mimura et al. (2004) have recently suggested on the basis of in vitro experiments that wild-type Cdc6 should not be able to reload during G2/M.
To address and disentangle these issues, we constructed versions of CDC6 in which expression in yeast could be well regulated independently of CDK activity, where expression of Cdc6 did not cause significant CDK inhibition and where Cdc6 expression and nuclear import were independent of CDK activity. We then asked whether phosphorylatable or nonphosphorylatable forms of Cdc6 forced into the nucleus in G2 phase could be reloaded onto chromatin while Clb-Cdc28 kinase was active; in a sense, these are in vivo versions of the in vitro experiments carried out by Mimura and coworkers. In agreement with Mimura et al. (2004), we found that phosphorylatable Cdc6 cannot be reloaded onto chromatin at moderate expression levels during G2/M, but nonphosphorylatable Cdc6 can be reloaded efficiently. This reloading does not on its own cause rereplication. However, in yeast cells where Cdc6 does reload, changes in other replication proteins can provoke rereplication.
MATERIALS AND METHODS
Plasmids and Strains
Yeast strains and plasmids are described in Tables 1 and 2. Genetic manipulations used standard techniques. CDC6-8A, ORC2-6A and ORC6-4A encode nonphosphorylatable mutant proteins in which alanine was substituted for serine or threonine in potential CDK phosphorylation sites at 7, 23, 39, 43, 135, 354, 367, and 372 for CDC6-8A; 16, 24, 70, 174, 188, and 206 for ORC2-6A; and 106, 116, 123, 146 for ORC6-4A. We refer to mutant CDC6-8A, ORC2- 6A, and ORC6-4A alleles as CDC6*, ORC2*, and ORC6*, respectively. The ORC2* and ORC6* alleles were derived from YJL1737 (A364a ORC2-6A ORC6-4A; Nguyen et al., 2001). Strains YSH82 (CDC6-HA3) and YSH120 (CDC6*-HA3) were constructed by transformation of YSH48 (W303a Δbar1:: LEU2) with CEN plasmids pSH33 (pCDC6-CDC6-HA3) or pSH40 (pCDC6-CDC6*-HA3), respectively. CDC6-HA3 on plasmid pSH33 was amplified by PCR using oligos PRS32 (CTGCTAGGATTACACATGGCATGGATGAACTATACAAAGGTGGTGGCATGTCAGCTATACCAATAACTCC) and PRS33 (AGTCATAGAAGCCATACCCACCTTGCGCTTTTTCTTTGGACCGCGGCCGCACTGAGCAGCGTAATC) to generate the CDC6-HA3 fragment flanked by sequences homologous to Sph1-cut linear plasmid pSH48. Plasmid pSH51N (pMET3-GFP-CDC6-HA3-NLS-T7-NLS) contained in YSH143N was constructed by homologous recombination between CDC6-HA3 fragment (above PCR product) and Sph1-cut linear pSH48 cotransformed into YSH140. Similarly, pSH53 (pMET3-GFP-CDC6*-HA3-NLS-T7-NLS) contained in YSH145 was constructed using CDC6*-HA3 fragment amplified from pSH40 with PRS34 (CTGCTAGGATTACACATGGCATGGATGAACTATACAAAGGTGGTGGCATGTCAGCTATACCAATAGCTCC) and PRS33. Constructs pMET3-GFP- CDC6-HA3-NLS-T7-NLS (pSH51N) and pMET3-GFP-CDC6*-HA3-NLS-T7-NLS (pSH53) are abbreviated as MET-CDC6-NLS and MET-CDC6*-NLS, respectively. Both CEN plasmids pSH51N and pSH53 were recovered from strains YSH143N and YSH145, respectively, and used to generate strains containing MET-CDC6-NLS or MET-CDC6*-NLS with various combination of ORC2* ORC6*, MCM7-2NLS, and MCM7-2NLS-3A (Table 1).
Table 1.
Yeast strains used in this study
| Name | Relevant genotypea | Backgroundb | Source |
|---|---|---|---|
| YSH82 | Δbar1::LEU2 [CDC6-HA3 URA3] | W | This study |
| YSH120 | Δbar1::LEU2 [CDC6*-HA3 URA3] | W | This study |
| YSH143N | Δbar1::HIS3 [pMET3-CDC6-NLS LEU2] | W | This study |
| YSH145 | Δbar1::HIS3 [pMET3-CDC6*-NLS LEU2] | W | This study |
| YSH140 | Δbar1::HIS3 | W | This study |
| YSH223 | Δbar1::HIS3 MCM7-NLS | W | This study |
| YSH224 | Δbar1::HIS3 MCM7-nls(3A) | W | This study |
| YJL1737 | MATa ade2 ade3 leu2 ura3-52trp1-289 his7 bar1::LEU2 ORC2-6A ORC6-4A | A | J. Li |
| YSH197 | bar1::TRP1 ORC2*ORC6* | A | This study |
| YSH199 | bar1::TRP1 ORC2*ORC6* MCM7-NLS | A | This study |
| YSH200 | bar1::TRP1 ORC2*ORC6* MCM7-nls(3A) | A | This study |
| YSH178N | Δbar1::HIS3 MCM7-NLS [pMET3-CDC6-NLS LEU2] | W | This study |
| YSH179N | Δbar1::HIS3 MCM7-nls(3A) [pMET3-CDC6-NLS LEU2] | W | This study |
| YSH201 | bar1::TRP1 ORC2*ORC6* [pMET3-CDC6-NLS LEU2] | A | This study |
| YSH207 | bar1::TRP1 ORC2*ORC6* MCM7-NLS [pMET3-CDC6-NLS LEU2] | A | This study |
| YSH211 | bar1::TRP1 ORC2*ORC6* MCM7-nls(3A) [pMET3-CDC6-NLS LEU2] | A | This study |
| YSH209 | bar1::TRP1 ORC2*ORC6* MCM7-NLS [pMET3-CDC6*-m1-NLS LEU2] | A | This study |
| YSH213 | bar1::TRP1 ORC2*ORC6* MCM7-nls(3A)[pMET3-CDC6*-m2-NLS LEU2] | A | This study |
| GZY45-15c | MATa ade2-1 his3-11,15 leu2–3,112trp1–1 ura3-1 can1-100 ssd1-d bar1 fkh1::LEU2 fkh2::HIS3(Disomic for chromosome 16) | W | Zhu et al. (2000) |
| YSH253 | bar1::LEU2 ORC2*ORC6* [pGAL1-PRI1 URA3] | A | This study |
| YSH266 | bar1::LEU2 ORC2*ORC6* [pGAL1-CDC45 URA3] | A | This study |
| YSH249 | bar1::LEU2 ORC2*ORC6* [pGAL1-DPB11 URA3] | A | This study |
| YSH272 | Δbar1::HIS3 [pMET3-CDC6*-NLS LEU2] [pGAL1-PRI1 URA3] | W | This study |
| YSH285 | Δbar1::HIS3 [pMET3-CDC6*-NLS LEU2] [pGAL1-CDC45 URA3] | W | This study |
| YSH268 | Δbar1::HIS3 [pMET3-CDC6*-NLS LEU2] [pGAL1-DPB11 URA3] | W | This study |
| YSH291 | Δbar1::LEU2 [pGAL1-PRI1 URA3] | W | This study |
| YSH304 | Δbar1::LEU2 [pGAL1-CDC45 URA3] | W | This study |
| YSH287 | Δbar1::LEU2 [pGAL1-DPB11 URA3] | W | This study |
| YSH307 | MATa ade2 ade3 leu2 ura3-52 trp1-289 his7 bar1::LEU2 [cir+]ORC2-6A ORC6-4A | A | This study |
| YSH310 | bar1::LEU2 [cir+]ORC2*ORC6* [pGAL1-DPB11 URA3] | A | This study |
| YSH61 | pGAL-CDC6–9XMYC URA3 | W | This study |
| YSH63 | pGAL-CDC6*-9XMYC URA3 | W | This study |
All strains contain a wild-type allele of CDC6 at the natural CDC6 locus. All strains are MATa.
a ORC2*: ORC2-6A; ORC6*: ORC6-4A; CDC6*: CDC6-8A. NLS and nls(3A) are two copies of the active and mutated (inactive) version of SV40 NLS, respectively.
b W, W303a (MATa ade2-1 his3-11,15 ura3-1 leu2-3,112 trp1-1 can1-100 ssd1-d [psi+]; A, A364a (MATa ade1 ade2 ura1 his7 lys2 tyr1 gal1). W303 is from R. Rothstein (Thomas and Rothstein, 1989), YJL1737 is from J. Li (Nguyen et al., 2001), and GZY45-15c is from Zhu et al. (2000).
Table 2.
Plasmids used in this study
| Name | Relevant genotype | Source |
|---|---|---|
| pSH33 | pAlter(CDC6-HA3 URA3) | G. Sherlock, unpublished data |
| pSH40 | pAlter(CDC6*-HA3 URA3) | G. Sherlock, unpublished data |
| pSH48 | pRS315(pMET3-GFP-Sph1-Not1-NLS-T7-NLS LEU2) | N. Edgington |
| pSH51N | pRS315(pMET3-GFP-CDC6-HA3-NLS-T7-NLS LEU2) | This study |
| pSH53 | pRS315(pMET3-GFP-CDC6*-HA3-NLS-T7-NLS LEU2) | This study |
| pJL1206 | (MCM7-2NLS URA3) | J. Li |
| pKI1260 | (MCM7-2NLS3A URA3) | J. Li |
| Name |
Relevant genotype |
ORF ID |
| pSH83 | BGK1805 (GAL1-MCM10 URA3 | YIL150C |
| pSH84 | BGK1805 (GAL1-DPB11 URA3) | YJL090C |
| pSH85 | BGK1805 (GAL1-SLD2 URA3) | YKL108W |
| pSH86 | BGK1805 (GAL1-NOC3 URA3) | YLR002C |
| pSH87 | BGK1805 (GAL1-POL12 URA3) | YBL035C |
| pSH88 | BGK1805 (GAL1-PRI1 URA3) | YIR008C |
| pSH89 | BGK1805 (GAL1-SLD3 URA3) | YGL113W |
| pSH90 | BGK1805 (GAL1-POL30 URA3) | YBR088C |
| pSH91 | BGK1805 (GAL1-DPB3 URA3) | YBR278W |
| pSH94 | BGK1805 (GAL1-SLD5 URA3) | YDR489W |
| pSH95 | BGK1805 (GAL1-PSF1 URA3) | YDR013W |
| pSH96 | BGK1805 (GAL1-PSF2 URA3) | YJL072C |
| pSH97 | BGK1805 (GAL1-PSF3 URA3) | YOL146W |
| pSH98 | BGK1805 (GAL1-DPB2 URA3) | YPR175W |
| pSH101 | BGK1805 (GAL1-CDC45 URA3) | YLR103C |
| pSH104 | BGK1805 (GAL1-MCM2 URA3) | YBL023C |
| pSH105 | BGK1805 (GAL1-MCM3 URA3) | YEL032W |
| pSH106 | BGK1805 (GAL1-MCM4 URA3) | YPR019W |
| pSH107 | BGK1805 (GAL1-MCM5 URA3) | YLR274W |
| pSH108 | BGK1805 (GAL1-MCM7 URA3) | YBR202W |
| pSH109 | BGK1805 (GAL1-MOB1 URA3) | YIL106W |
| pSH113 | BGK1805 (GAL1-BUD4 URA3) | YJR092W |
Plasmids from N. Edgington are described in Edgington and Futcher (2001); plasmids from J. Li are described in Nguyen et al. (2001). pSH83 through pSH113 were prepared from a yeast ORF clone (in E. coli) purchased from Open BioSystems (Huntsville, AL).
pJL1206 (MCM7-2NLS) and pKI1260 (MCM7-2NLS3A) (from J. Li; Nguyen et al., 2001) encode Mcm7 protein fused at its C-terminus to two copies of the active and mutated (inactive) version of SV40 NLS, respectively. These integrating plasmids were used to replace the resident MCM7 gene with MCM7-2NLS or MCM7-2NLS3A (abbreviated as MCM7-NLS and MCM7-nls(3A), respectively).
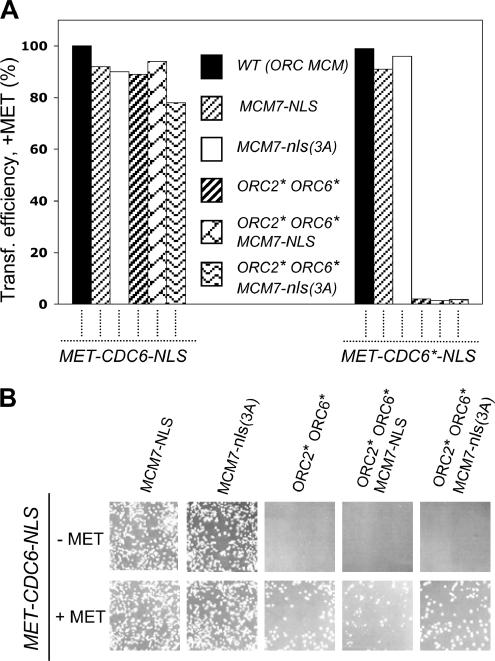
Strains in Figure 5 were constructed by placing MET-CDC6-NLS (pSH51N) and MET-CDC6*-NLS (pSH53) individually in the following strains: YSH140 (WT), YSH223 (MCM7-NLS), YSH224 [MCM7-nls(3A)], YSH197 (ORC2* ORC6*), YSH199 (ORC2* ORC6* MCM7-NLS), and YSH200 [ORC2* ORC6* MCM7-nls(3A)].
Figure 5.
Cdc6* is extremely toxic in ORC2* ORC6* strains. (A) MET-CDC6-NLS (pSH51N) and MET-CDC6*-NLS (pSH53) were individually transformed into strains of the indicated genotypes [left to right, YSH140, WT; YSH223, MCM7-NLS; YSH224, MCM7-nls-(3A); YSH197, ORC2* ORC6*; YSH199, ORC2* ORC6* MCM7-NLS; YSH200, ORC2* ORC6* MCM7-nls(3A); nls(3A) is an inactive version of the NLS]. Cells were spread on +met −leu plates (to repress the MET promoter and to select for the plasmid). The number of colonies was counted after 3 d at 30°C. For each plasmid, the number of transformants was normalized to the number of transformants obtained in the wild-type strain, which was several thousand. Similar results were obtained in multiple independent experiments. (B) Cells containing MET-CDC6-NLS (pSH51N) were grown to log phase in a medium containing 2 mM methionine. After washing, cells were spread on either +met or −met plates, and these plates were incubated for 3 d at 30°C and photographed.
A DDC2-GFP kanMX6 construct was made by PCR using genomic DNA from strain yJK7-2 (Melo et al., 2001) along with oligonucleotides PRS72 (AAAGGTACGTGGGACAAGAC) and PRS73 (AGACAGCAACACACATCTAG). Yeast strains containing DDC2-GFP kanMX indicated in Table 5 were generated by replacing the DDC2 locus with above purified PCR product via homologous recombination. Transformants were selected on G418-plates.
Table 5.
Ddc2-GFP foci
| Genotype | +DNA? | Toxic? | Foci/100 cells |
No. of foci/cell |
||
|---|---|---|---|---|---|---|
| Suc | Induced | 1 | >1 | |||
| ORC* MET-CDC6 MCM7-NLS | Yes | Yes | 7 | 23 | 17 | 6 |
| ORC* MET-CDC6 MCM7-NLS | Yes | Yes | 6 | 38 | 25 | 13 |
| ORC* MET-CDC6 MCM7-NLS(3A) | Yes | Yes | 6 | 13 | 11 | 2 |
| ORC* [cir+] | No | No | 5 | 5 | 5 | 0 |
| CDC6* | No | No | 4 | 5 | 5 | 0 |
| ORC* GAL-DBP11 | Yes | Yes | 8 | 15 | 13 | 2 |
| CDC6* GAL-DBP11 | Yes | Yes | 10 | 17 | 14 | 3 |
| WT GAL-DBP11 | No | No | 4 | 5 | 5 | 0 |
| ORC* GAL-PRI1 | No | No | 5 | 4 | 4 | 0 |
| CDC6* GAL-PRI1 | No | No | 5 | 6 | 6 | 0 |
| WT GAL-PRI1 | No | No | 4 | 4 | 4 | 0 |
In addition to the indicated genotype, all strains contain DDC2-GFP-kanMX integrated at their DDC2 locus. ORC* means ORC2* ORC6*. +DNA? is Yes if a shift to >2N DNA content is seen by flow cytometry. Toxic? is Yes if the strain grows poorly upon induction of the conditional gene. The number of cells with at least one Ddc2-GFP focus, per 100 cells, is shown without induction (Suc) or with induction of the conditional gene (+Gal, or −Met, or both, as the case may be). By a Chi-squared test, the galactose-induced ORC* GAL-DBP11 and CDC6* GAL-DBP11 strains have significantly more cells with foci than their negative controls (p < 0.001). Induced cells with at least one Ddc2-GFP focus are classified into cells that have exactly one focus or two or more foci.
Chromatin-associated Protein Analysis
Chromatin-associated proteins were analyzed as described (Liang and Stillman, 1997) with modifications. Fractionated cells were incubated in prespheroplasting buffer for 15 min on ice before spheroplasting in 50 μl of 1.5 mg/ml Oxalyticase (Enzogenetics, Corvallis, OR). Spheroplasts were washed twice with lysis buffer before lysing in the presence of 1% Triton X-100. The quality of chromatin-pellet fraction was monitored by checking the presence of chromain-bound Orc3 and absence of cytosolic Adh using monoclonal anti-Orc3 (SB3) and rabbit polyclonal anti-Adh. Primary antibodies used for immunoblot analysis of proteins separated on 10% SDS-PAGE were as follows: 12CA5 anti-HA monoclonal ascites, 9H8/5 anti-Cdc6 mouse monoclonal ascites, and SB3 anti-Orc3 mouse monoclonal ascites. Chemiluminescence signal on immunoblots was detected using Supersignal reagents (Pierce, Rockford, IL).
Fluorescence Microscopy and Fluorescence-activated Cell Sorting Analysis
MET-GFP-CDC6-NLS and MET-GFP-CDC6*-NLS cells were grown in medium containing 140 μM methionine and examined using differential interference contrast for morphological analysis or fluorescence microscopy to visualize green fluorescent protein (GFP) or 4,6-diamidino-2-phenylindole (DAPI). DAPI staining was performed on cells fixed typically for 1–2 h in formaldehyde. In Figure 3, fixation of cells in formaldehyde was for 10 min or less to visualize both GFP and DAPI in the same cell. Images were obtained with a Zeiss AxioCam camera (Thornwood, NY) mounted on an Olympus BH-2 microscope (Melville, NY) and captured using Openlab 3.0.1 software from Improvision (Lexington, MA). DAPI and GFP images were pseudocolored and digitally merged using the Openlab 3.0.1 software. For fluorescence-activated cell sorting (FACS) analysis, cells were stained with propidium iodide.
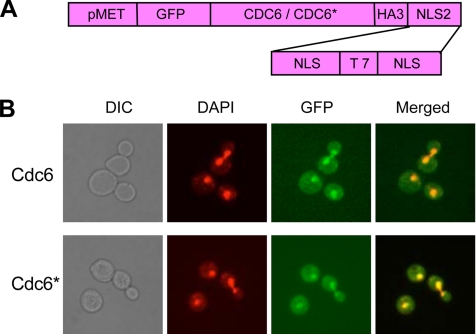
Figure 3.
MET-CDC6-NLS and MET-CDC6*-NLS encode proteins constitutively present in the nucleus at all cell cycle stages. (A) The structures of MET-GFP-CDC6-HA3-NLS2 and MET-GFP-CDC6*-HA3-NLS2. These constructs are referred to as “MET-CDC6-NLS” and “MET-CDC6*-NLS.” (B) Strains YSH143N (MET-CDC6-NLS) and YSH145 (MET-CDC*6-NLS) were grown in medium containing 140 μM methionine (a partially repressing condition). Cells were stained with DAPI and then examined by fluorescence and bright-field microscopy.
Viability and Rereplication Assays
Log phase cells in medium containing 2 mM methionine were arrested with α-factor (60 nM). After 2 h, the α-factor and methionine were removed by washing. The arrested cells were then resuspended in fresh medium containing 15 μg/ml nocodazole and no methionine. Samples were taken hourly for DNA and viability assays. For viability assays, 500 cells were counted using Coulter counter (Beckman, Fullerton, CA) at the 0-hr sample, and the same volume of sonicated cells at the 3-h time-point sample was plated on +MET plates (containing 2 mM methionine) and −MET plates (containing no methionine). Colonies were counted after 3 d at 30°C.
Microarrays
Genomic DNA was isolated using Genomic DNA Buffer Set (Qiagen, Chatsworth, CA). Isolated genomic DNAs were purified using Qiagen Genomic-tips. Labeled cDNA probe was synthesized from 4 μg purified genomic DNA incubated with 240 μM aminoallyl-dUTP (aadUTP), 200 ng/μl random hexamer, 360 μM dNTPs, and 120 μM dTTP using Klenow fragment at 37°C for 4–5 h. The resulting aadUTP-cDNA probe was purified using the Qiagen PCR purification kit and coupled with Cy3 or Cy5 fluorescent dye using a protocol from TIGR (http://www.tigr.org/tdb/microarray/protocolsTIGR.shtml). Purified coupled cDNAs corresponding to 100 pmol Cy3 and 100 pmol Cy5 were mixed together and hybridized, as described by Oliva et al. (2005), to microarrays printed by spotting PCR products onto glass slides coated with amino-propylsilane. These microarrays were exactly as described (Oliva et al., 2005), but with S. cerevisiae PCR fragments.
Analysis of Ddc2-GFPp Foci
Logarithmically growing cells in medium containing sucrose were washed and divided into two halves. One-half of the cells were reconstituted in medium with sucrose (2%), and galactose was added to 2% to the other half of cells. After a 4-h incubation at 30°C, cells were visualized on an Olympus BH-2 microscope, and images were recorded using Openlab 3.0.1 software. The number of foci (0, 1 or 2, or more than 2) per cell was quantified for 100–150 cells from each strain growing in the presence of sucrose or galactose. To induce expression of CDC6* cells were grown in medium lacking methionine.
RESULTS
Cdc6*, But Not Cdc6, Reloads onto Chromatin at a Nocodazole Block
The Cdc6 protein has eight occurrences of Ser-Pro (SP) or Thr-Pro (TP) (potential phosphorylation sites for Cdc28 kinase) including six perfect Cdc28 consensus sites |(S/T)-P-X-(K/R)}. We mutated all eight potential phospho-acceptors to alanine and generated a nonphosphorylatable mutant called CDC6*. Strains containing CDC6* as the only source of Cdc6 do not have discernable growth defects (Sherlock and Futcher, unpublished data) consistent with previous results (Nguyen et al., 2001).
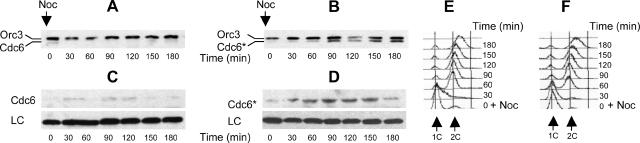
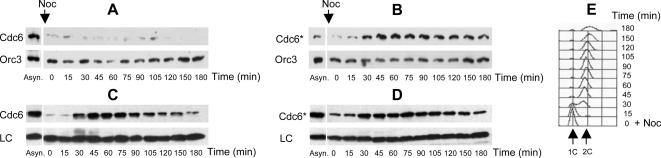
If Cdc6 fails to reload onto chromatin in G2/M solely because of direct effects of CDK activity, then Cdc6* ought to be able to reload. To see if Cdc6* does reload, Cdc6 and Cdc6* proteins were tagged with the HA epitope (Cdc6-HA3 and Cdc6*-HA3) and expressed from the endogenous CDC6 promoter. Cells were arrested in G1 with α-factor. Arrested cells were then released into medium containing nocodazole, and cells then arrested in G2/M phase with high Clb-Cdc28 kinase activity. The majority of cells replicated their DNA by 60 min (Figure 2, E and F) and these remained arrested in G2/M as large budded cells (data not shown) for the duration of the experiment. Cdc6*, but not Cdc6, reloaded onto chromatin 90 min after addition of nocodazole (Figure 2). Thus, the nonphosphorylatable form of Cdc6, but not the wild-type form, can reassociate with chromatin, whereas Clb-Cdc28 kinases are active, suggesting that phosphorylation of Cdc6 is indeed a major control on reloading in vivo, by one mechanism or another.
Figure 2.
Cdc6*, but not Cdc6, reloads onto chromatin at a nocodazole block. Cells of strain YSH82 (CDC6-HA3) (A, C, and E) or YSH120 (CDC6*-HA3) (B, D, and F) were arrested in G1 with α-factor. Arrested cells were released (time 0) and nocodazole (Noc; 15 μg/ml) was added to arrest cells in G2/M. Samples were taken every 30 min. Samples were processed to yield crude chromatin pellets (A and B; see Materials and Methods) or whole cell extracts (C and D). (A and B) Chromatin-associated proteins were assayed for Cdc6 (A) or Cdc6* (B) by immunoblotting with anti-HA antibody. Orc3 was used as a loading control. (C and D) Whole cell extracts from the same samples as A and B were assayed for total Cdc6 (C) or Cdc6* (D) by immunoblotting with anti-HA antibody. A cross-reacting band of unknown origin was used as a loading control (LC). (E and F) Flow cytometry was used to assay cell cycle position; 1C and 2C DNA peaks are indicated.
Construction of Strains Constitutively Expressing Cdc6 or Cdc6* in the Nucleus
There are several explanations for the inability of wild-type Cdc6 to reload: first, the protein might be absent, because of phosphorylation-induced degradation; this seems to be part of the explanation, as shown by Western analysis (Figure 2). But additional mechanisms might also exist. The protein might be phosphorylated and retained in the cytoplasm due to an ineffective nuclear localization signal, or phosphorylation of Cdc6 might directly or indirectly block its association with chromatin. To examine these possibilities, we constructed versions of Cdc6 and Cdc6* that allowed us to control localization and expression. We added two copies of the SV40 nuclear localization signal (NLS), which directs proteins to the yeast nucleus (Edgington and Futcher, 2001), to the C-termini of Cdc6 and Cdc6*. We also made control constructs with mutant, nonfunctional NLSs. We tagged the N-termini of these proteins with GFP, so that the amount and location of each protein could be assayed. Each gene was cloned behind the repressible MET3 promoter, allowing regulation of expression using different amounts of methionine (Figure 3A).
Expression of these constructs (MET3-GFP-CDC6-HA3-NLS2 and MET3-GFP-CDC6*-HA3-NLS2, hereafter called MET-CDC6-NLS, and MET-CDC6*-NLS) from the MET promoter in the presence of 140 μM methionine (a partially repressing condition) complemented a cdc6-ts mutant at restrictive temperature, showing that the tagged proteins are functional. Fluorescence microscopy showed constitutive expression and predominant nuclear localization of both Cdc6 and Cdc6* fusion proteins throughout the cell cycle (Figure 3B). When MET-CDC6 is expressed in 140 μM methionine, and MET-CDC6* (encoding a more stable protein) is expressed in 170 μM methionine, the amounts of Cdc6 and Cdc6* proteins are similar to each other and similar to wild-type amounts of Cdc6 (Figures 3 and 4 and data not shown). Furthermore, these levels of Cdc6 and Cdc6* did not cause the abnormal, elongated bud morphology seen when Cdc6 is overexpressed (Elsasser et al., 1996); the abnormal buds are due to inhibition of CDK activity by excess Cdc6, and the absence of abnormal buds suggests that levels of Cdc6 are indeed close to wild type and not high enough to cause significant CDK inhibition. We note that on 0 mM methionine, where expression is higher, MET-CDC6 does cause most cells to have the elongated bud morphology typical of CDK inhibition. On 0 mM methionine, MET-CDC6* causes only a few percent of the cells to have elongated buds; Cdc6* is known to be a relatively poor CDK inhibitor (Mimura et al., 2004).
Figure 4.
Cdc6*-NLS loads onto chromatin when Cdc6-NLS does not. MET-CDC6-NLS (i.e., MET-GFP-CDC6-HA3-NLS2, YSH143N) and MET-CDC6*-NLS (i.e., MET-GFP-CDC6*-HA3-NLS2, YSH145) were constitutively expressed by growing cells in medium containing 140 or 170 μM methionine, respectively. Cells were arrested in G1 with α-factor for 2 h. Cells were released from G1 at time 0, and nocodazole (Noc; 15 μg/ml) was added to arrest cells in G2/M. Samples were taken in exponential growth (Asy) and at 15-min intervals after release from α-factor. Samples were processed to yield crude chromatin pellets (A and B; Materials and Methods) or whole cell extracts (C and D). (A and B) Chromatin-associated proteins were assayed for Cdc6 (A) or Cdc6* (B) by immunoblotting with anti-HA antibody. Orc3 was used as a loading control. (C and D) Whole cell extracts from the same samples as A and B were assayed for total Cdc6 (C) or Cdc6* (D) by immunoblotting with anti-HA antibody. A cross-reacting band of unknown origin was used as a loading control (LC). (E) Flow cytometry showed that S-phase was largely completed by 30 min in the YSH145 (CDC6*) strain. Virtually identical results were obtained with YSH143N (CDC6). Fluorescence microscopy showed that in both strains, Cdc6 was present in the nucleus at the nocodazole arrest (not shown).
Cdc6*-NLS Loads onto Chromatin When Cdc6-NLS Does Not
To see whether constitutive moderate expression and nuclear localization allow Cdc6 to load onto chromatin in the presence of active Clb-Cdc28, we performed an α-factor release, nocodazole block experiment similar to that shown in Figure 2. Strains YSH143-N (MET-CDC6-NLS) and YSH145 (MET-CDC6*-NLS) were grown in medium containing partially repressing levels of methionine, arrested in G1 with α-factor, and then released into medium containing nocodazole. Most cells replicated DNA by 30–45 min after release from α-factor (Figure 4), and more than 90% of the cells then accumulated at the nocodazole block with a 2C DNA content. Chromatin precipitation was used to assay the amount of Cdc6 or Cdc6* associated with chromatin at various times. In the initial, asynchronous cells, both Cdc6 and Cdc6* were present on the chromatin. After 2 h in α-factor, both Cdc6 and Cdc6* had been released from chromatin, having presumably already loaded Mcms. It appears that neither Cdc6 nor Cdc6* are able to reload onto the chromatin during this period (i.e., late G1 and S). Immediately after DNA synthesis, Cdc6* reloaded onto chromatin, but Cdc6 did not (Figure 4).
These data suggest that neither turnover nor localization of wild-type Cdc6 can fully account for its inability to reload onto chromatin in the presence of Clb-Cdc28 activity. Instead, it appears that the phosphorylation sites of Cdc6 act by some additional mechanisms to block reassociation with chromatin. This likely involves association with Clb2-Cdc28 (Mimura et al., 2004). Consistent with the results of Mimura et al. (2004) and Wolf et al. (1999), we find that Cdc28 is found in association with Cdc6, but not Cdc6* (Supplementary Figure 10).
Cdc6*-NLS Is Toxic and Causes Rereplication in ORC2* ORC6* Strains
The loading of Cdc6 onto chromatin is a critical step in the formation of a pre-RC (Liang et al., 1995; Cocker et al., 1996; Detweiler and Li, 1997). Thus, because Cdc6* reloads prematurely, strains expressing Cdc6* might be prone to rereplication. Expression of Cdc6* is not sufficient for rereplication, because our MET-CDC6* strains are healthy and do not show any abnormalities by flow cytometry. Nevertheless, Cdc6* might sensitize strains to rereplication.
Indeed, it has previously been shown (Nguyen et al., 2001; Vas et al., 2001; Mimura et al., 2004; Wilmes et al., 2004) that CDC6 mutants similar to CDC6* sensitize strains to rereplication. We looked for rereplication using our own CDC6 constructs, which are somewhat different from used previously. Our CDC6 alleles are nonphosphorylatable because of point mutations at the phosphorylation sites; they are constitutively nuclear because of the appended NLS; and they are expressed from the repressible MET promoter. In contrast, some previous experiments have been done with mutant proteins lacking amino acids 2–46; these proteins lack the N-terminal sequences for Cdc6 degradation (Drury et al., 1997; Elsasser et al., 1999), for association with Clb2/Cdc28 (Elsasser et al., 1996; Mimura et al., 2004), and for nuclear localization (Jong et al., 1996; Luo et al., 2003). Because these proteins lack the native NLS, they may have difficulty accessing the nucleus. Furthermore, the CDC6 allele of Nguyen et al. was overexpressed from the GAL promoter.
We obtained an ORC2* ORC6* strain containing a wild-type allele of CDC6 (YJL1737) from J. Li (Nguyen et al., 2001). We transformed MET-CDC6-NLS or MET-CDC6*-NLS into this strain, with or without an MCM7-NLS/nls(3A) plasmid (Nguyen et al., 2001). Partial results are shown in Figure 5. Two interesting findings were, first, that even though the transformations were spread onto plates containing 2 mM methionine to repress the MET promoter, we were unable to place the MET-CDC6*-NLS plasmid into any strain that also contained the ORC2* ORC6* mutations (whether MCM7-NLS was present or not). In an otherwise wild-type strain, the Cdc6* expressed from the MET promoter on 2 mM methionine is undetectable by Western blotting; nevertheless, we believe that MET-CDC6*-NLS is expressed at a low level on 2 mM methionine and that in the presence of ORC2* ORC6*, it is extremely toxic, presumably because of rereplication (see below). In contrast, MET-CDC6*-NLS efficiently transformed strains with wild-type ORC genes, and these strains had no significant phenotype with or without methionine, or with or without MCM7-NLS.
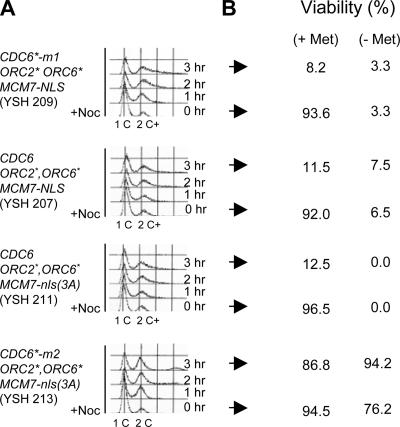
Second, with MET-CDC6-NLS, we obtained ORC2* ORC6* transformants, but only on plates containing methionine. In the absence of met (i.e., when MET-CDC6 was expressed), few or no transformants were obtained. This suggests that moderate overexpression of Cdc6-NLS in the presence of ORC2* ORC6* causes rereplication. Again, presence or absence of MCM7-NLS made no difference. When a MET-CDC6-NLS ORC2* ORC6* strain is grown in the absence of methionine, it arrests and accumulates cells with slightly more than 2C DNA content (see Figure 6 for a related example), presumably due to rereplication. Even a brief absence of methionine is lethal to these strains (Figure 6).
Figure 6.
Excess DNA from rereplication is correlated with toxicity. Strains YSH209 [MET-CDC6*-m1-NLS ORC2* ORC6* MCM7-NLS), YSH207 (MET-CDC6-NLS ORC2* ORC6* MCM7-NLS), YSH211 (MET-CDC6-NLS ORC2* ORC6* MCM7-nls(3A), and YSH213 (MET-CDC6*-m2-NLS ORC2* ORC6* MCM7-nls(3A)] were grown in +met medium to repress MET-CDC6/CDC6*. Cells were arrested with α-factor and then released into −met medium with 15 μg/ml nocodazole at 0 time. Cells were sampled every hour and DNA content and viability were assayed. (A) Flow cytometry. In the CDC6*-m1 strain and the two CDC6 strains (i.e., YSH209, 207, and 211), a 2C peak appears at the nocodazole arrest, and this peak drifts to the right, signifying DNA content slightly higher than 2C. In contrast, the peak in the CDC6*-m2 strain (YSH213) remains at 2C. (B) Viability. After 0 or 3 h without methionine, cells were sampled and spread on −met or +met plates, and viability was assayed. For the three strains showing more than 2C DNA (i.e., YSH209, 207, and 211), after 3 h of exposure to −met medium, only 8–13% of the cells could be rescued by spreading back onto +met plates. In contrast, the CDC6*-m2 cells remained viable with or without methionine.
Thus, both Cdc6-NLS and Cdc6*-NLS are toxic to an ORC2* ORC6* strain, but Cdc6*-NLS is toxic even at extremely low levels, undetectable by Western blotting, whereas Cdc6-NLS is toxic only when expressed at higher levels. Overexpressed Cdc6 acts as a CDK inhibitor (Calzada et al., 2001), and so we presume that it allows accumulation of some unphosphorylated Cdc6, which then reloads and causes rereplication, just as if it were Cdc6* (see Discussion).
The MET-CDC6-NLS construct can transform an otherwise wild-type strain even when the MET promoter is turned on; so again, the ORC2* ORC6* mutations seem to be needed for rereplication. With the MET promoter turned on, MET-CDC6-NLS ORC2 ORC6 cells show abnormal, elongated buds, which we believe indicate CDK inhibition due to Cdc6 overexpression (Elsasser et al., 1996).
Rare transformants with heterogeneous phenotypes were obtained from the MET-CDC6*-NLS plasmid in ORC2* ORC6* strains on +met plates (Figures 5 and 6). Initially we were concerned that these might represent rare but real transformants. However, when the transforming plasmid from these rare clones was passaged through Escherichia coli and retransformed into yeast, we found that all recovered transforming plasmids gave thousands of transformants (compared with 0–5 transformants in parallel transformations with the original plasmid), suggesting that all these plasmids contained attenuating mutations. Yeast cells from the rare transformed clones were cured of their plasmid and retransformed with the original plasmid, and again 0–5 transformants were obtained (compared with thousands of transformants with a control plasmid lacking a CDC6* insert), suggesting that the rare transformants were not due to mutations in the yeast cells.
Further evidence that that the transformants were due to extra mutations in the CDC6* on the plasmid was obtained by partial characterization of some of these plasmids. The transformants fell into two classes. The smaller class consisted of transformants that grew on +met plates, but died on −met plates (i.e., conditional dominant lethal phenotype). One such rare transformant was strain YSH209 (MET-CDC6*-m1-NLS ORC2* ORC6* MCM7-NLS); we call this CDC6* allele MET-CDC6*-m1-NLS (or CDC6*-m1). This transformant was sick and slow-growing even on 2 mM met plates. When shifted to −met medium, essentially all plasmid-bearing cells died, and flow cytometry showed a slight but distinct shift to higher DNA content (Figure 6). The toxicity is irreversible; once MET-CDC6*-m1-NLS is expressed at a significant level, cell death follows even if expression is quickly rerepressed. The shift to higher DNA content, and the irreversibility of the toxicity, suggest that these cells undergo DNA rereplication.
The larger class consisted of transformants that grew equally well on +met and −met plates (consistent with the idea that their plasmids contained null alleles of CDC6*). One such rare transformant contains an allele we call MET-CDC6*-m2-nls (YSH213, Figure 6, bottom panel). CDC6*-m2 causes neither toxicity nor rereplication. For all transformants examined, the shift to higher DNA content correlates perfectly with loss of viability after brief de-repression of the MET promoter.
To further characterize CDC6*-m1 and m2, we sequenced these two alleles, from the MET promoter through GFP and the CDC6* open reading frame and into the 3′ UTR. CDC6*-m2 had a −1 frameshift mutation at base 833 of CDC6*, shifting the reading frame of the second half of the protein. In addition, the “A” normally found at position 835 was mutated to a “C.” That is, the wild-type sequence TGGACAGAG was replaced by the mutant sequence TGGCCGAG. Because of the frameshift, CDC6*-m2 is almost certainly a null mutation, consistent with its phenotype.
CDC6*-m1 had a single base change from T to G at position 1154 (i.e., AAA ATA GGC became AAA AGA GGC). This changes codon 385 from isoleucine to arginine. Codon 385 is conserved in fungi, in that the residue at this position in 21 sequenced fungi is I, V, or A, and furthermore it is found in a fungally conserved stretch of amino acids. Thus it is plausible that the nonconservative change from I to R may cause a significant change in the function of the protein. This I to R change occurs near the beginning of the “winged helix” or “forkhead” DNA-binding domain found in the C-terminal third of Cdc6, and thus the mutation could affect DNA binding. Because MET-CDC6* is a dominant lethal in an ORC2* ORC6* strain (even under +met conditions), we view CDC6*-m1 as a likely attenuated or hypomorphic allele, possibly because it binds DNA less well.
Microarray Analysis of Rereplication
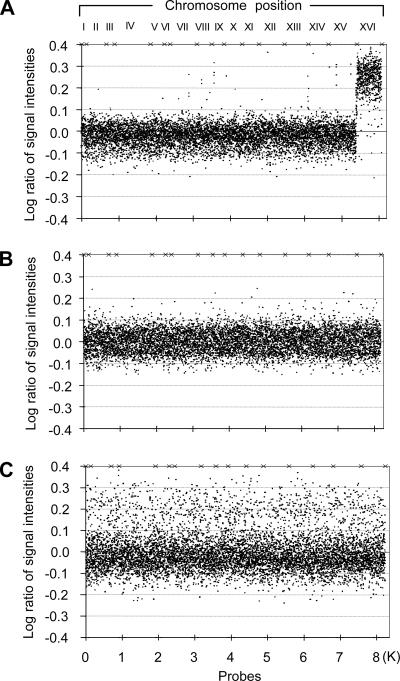
We used microarrays to analyze the extent of rereplication. DNA was extracted from a rereplicating strain, a wild-type control, or from a chromosome 16 disome, and labeled with fluorescent dye. DNA from a wild-type strain was labeled with a second fluorescent dye. The labeled DNAs were mixed and hybridized to a DNA microarray to determine relative DNA copy number at each probe on the array. In the WT control, copy numbers centered at 1 (Figure 7), as expected. In the chromosome 16 disome, copy numbers centered at 1 for chromosomes 1–15, but centered at 2 for chromosome 16 (Figure 7), showing that these microarrays are capable of detecting a twofold difference in copy number. For the rereplicating strain, and unlike the control strain, many DNA probes showed copy numbers between 1 and 2. Scatter around a copy number of 1 is asymmetric, with many more probes showing copy numbers significantly higher than 1 than lower than 1. Similar asymmetric scatter, with many probes showing copy numbers higher than 1, was seen in all three experiments done with the rereplicating strain, including one dye-swap experiment. The resolution of this experiment in terms of chromosomal position was relatively low compared with other recent microarray studies of rereplication (Green et al., 2006; Tanny et al., 2006), but the many probes showing copy numbers higher than 1 support the idea that rereplication is occurring in these strains.
Figure 7.
Microarray analysis of a rereplicating strain. DNA from control or experimental strains was extracted and fluorescently labeled with Cy3 and mixed with Cy5-labeled DNA from a wild-type control strain (for normalization). These mixtures were hybridized to spotted DNA microarrays. The log of the ratio of signal intensities (intensity in control or experimental strain divided by intensity in wild-type normalization strain) is plotted against chromosome position. A log ratio of 0.0 indicates a relative DNA copy number of 1, whereas a log ratio of 0.3 indicates a relative DNA copy number of 2. (A) Disomic strain. A strain disomic for chromosome 16 (GZY45–15c) was grown to log phase, and DNA was extracted and fluorescently labeled with Cy3. (B) Control strain. A nonrereplicating strain was arrested with nocodazole, and expression of MET-CDC6 was induced by removal of methionine. DNA was extracted and labeled with Cy3. (C) Rereplicating strain. A rereplicating strain (YSH209, see Figure 6) was arrested with α-factor, and released into a nocodazole block with removal of methionine (to induce MET-CDC6*-m1-NLS). After 3 h, DNA was extracted and labeled with Cy3.
ORC2* ORC6* and CDC6* Strains Are Sensitive to the Overexpression of Other Replication Proteins
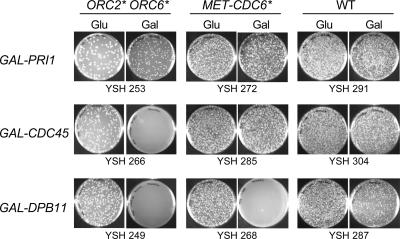
Although ORC2* ORC6* strains and CDC6* strains are individually quite healthy, the combination is lethal. This synergistic interaction might suggest there are two pathways for preventing rereplication: one represented by the Orc2/Orc6 and the other represented by Cdc6. If this is the correct view, then it might be that ORC2* ORC6* mutations would be synthetically lethal with various perturbations of the “Cdc6 pathway,” and similarly, CDC6* mutations would be synthetically lethal with perturbations of the “Orc2 Orc6 pathway.” To test this idea, we chose a panel of replication proteins cloned behind the GAL promoter and overexpressed them in a wild-type strain, a CDC6* strain, an ORC2* ORC6* strain, and an ORC2* ORC6* strain that lacked its native 2-μm circle plasmid (and therefore, because of poor partition at mitosis, carried the 2-μm–based GAL overexpression plasmids at very high copy numbers). In a first experiment, we spread the various transformants on galactose plates, and asked whether the strains were viable or not. Results are shown in Table 3.
Table 3.
Toxicity of replication proteins overexpressed in ORC2* ORC6* or CDC6* cells
| Description | WT | CDC6* | ORC* [ciro] | ORC* [cir+] | |
|---|---|---|---|---|---|
| MCM10 | At Ori; interacts Mcms | s s s s | m m m m | d d d d | d d d |
| DPB11 | At Ori; interacts Sld2 | + + | m m m + | d d d d | s s s |
| SLD2 | At Ori; interacts Dpb11 | + + + + | s s s s | s s s + | s s s |
| NOC3 | At Ori | + + + + | s s s s | d s s s | m m m |
| SLD3 | At Ori; interacts Cdc45 | + + + + | + + + + | d m m m | s s s |
| PSF1 | Subunit GINS | + + + + | + + + + | m m m m | s s s |
| PSF3 | Subunit GINS | + + + + | + + + + | s s s s | s s s |
| CDC45 | At Ori; interacts Sld3 | + + + + | + + + + | m m d d | s s s |
| POL12 | Subunit of Primase | + + + + | + + + + | + + + + | |
| PRI1 | Subunit of Primase | + + + + | + + + + | + + + + | + + + |
| POL30 | PCNA; sliding clamp | + + + + | + + + + | + + + + | |
| DPB2 | Subunit DNA Pol II | + + + + | + + + + | + + + + | |
| DPB3 | Subunit DNA Pol II | + + + + | + + + + | + + + + | |
| SLD5 | Subunit GINS | + + + + | + + + + | + + + + | |
| PSF2 | Subunit GINS | + + + + | + + + + | + + + + | |
| MCM2 | Subunit Mcm complex | + + + | + + + | + + + | |
| MCM3 | Subunit Mcm complex | m m m | m m m | m m m | |
| MCM4 | Subunit Mcm complex | + + + | + + + | + + + | |
| MCM5 | Subunit Mcm complex | + + + | + + + | + + + | |
| MCM7 | Subunit Mcm complex | m m m | m m m | m m m | |
| BUD4 | Bud-site selection; CDK sites | s s m | s s s | s s s | |
| MOB1 | Mitotic exit; CDK sites | + + + | + + + | + + + |
Results were assayed after 3 d on galactose plates at 30°C. In most cases, four independent transformants were tested; colony growth was scored as follows: +, normal growth; s, small and visible to the eye (typically more than 1000 cells); m, micro-colony (typically <1000 cells); d, dead and no visible growth. Bud4 is a negative control containing multiple CDK phosphorylation sites and causing moderate toxicity even in wild-type cells. Mob1 is a negative control containing multiple CDK phosphorylation sites and causing no toxicity.
There are several points of interest. First, proteins primarily involved in the elongation step of DNA synthesis (subunits of primase, PCNA, subunits of DNA polymerase) generally cause no toxicity (Table 3; though the relatively poorly understood proteins Psf1 and Psf3 could be an exception to this). Second, proteins that act at the origin to promote initiation often do cause toxicity (Cdc45, Sld2, Sld3, Dpb11; Figure 8, Table 3). Third, higher copy number causes higher toxicity, as expected (compare the ORC2* ORC6* strains with and without native 2-μm circle). Fourth, and most important to the idea we are testing, there is little or no complementarity to the toxicity. Instead, the proteins toxic to a CDC6* strain are a subset of the proteins toxic to an ORC2* ORC6* strain. This partial similarity in the pattern of sensitivity suggests that the CDC6* mutant is sensitized to the overexpression of these initiator proteins in a qualitatively way similar to that of the ORC2* ORC6* mutant.
Figure 8.
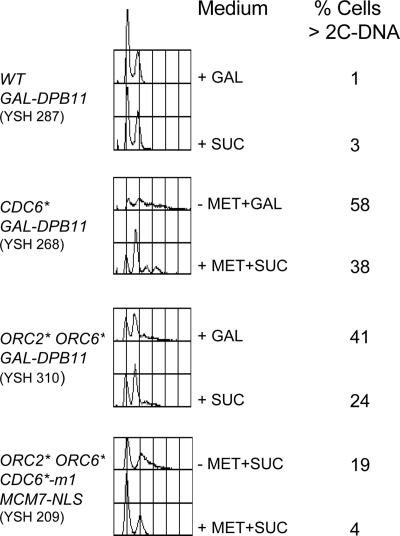
ORC2* ORC6* and CDC6* cells are sensitive to overexpression of replication initiation proteins. ORC2* ORC6* (YSH253, YSH266, and YSH249), MET-CDC6* (YSH272, YSH285, and YSH268), and WT (YSH291, YSH304, and YSH287) strains containing GAL-PRI1, GAL-CDC45, and GAL-DPB11 were grown to log phase in medium containing sucrose or sucrose plus 2 mM methionine for MET-CDC6* cells (to repress MET promoter). After washing three times with water, equal culture volumes of ORC2* ORC6* and WT cells were spread on galactose (to induce expression of PRI1, CDC45, and DPB11 from the GAL promoter) and glucose plates. MET-CDC6* cells were spread on glucose and galactose plates lacking methionine to activate CDC6* expression from the MET promoter. Plates were photographed after 3 d at 30°C.
Is the toxicity seen in Table 3 caused, at least in part, by rereplication? To find out, we did a viability experiment with the ORC2* ORC6*, or CDC6*, or the wild-type control strain containing GAL-DPB11, or GAL-SLD2, or GAL-PRI1 (as a control). Cells were grown to exponential phase in sucrose medium (i.e., GAL promoter off), and then the cells were switched (or not, as a control) to galactose medium for 4 h. Equal culture volumes were taken from the galactose cultures after 0 h (i.e., just before addition of galactose) or after 4 h, and spread on glucose plates. In this plating assay, the ORC2* ORC6* and the CDC6* strains containing GAL-DBP11 or GAL-SLD2 showed a slight decrease (∼5%) in the number of viable cells in the culture after 4 h in galactose despite an increase in the total number of cells as determined by cell counts. In contrast, all other strains showed an increase in viable cells in proportion to the increase in cell number (Table 4). This suggests that the toxicity is irreversible in at least some of the cells, and this supports the idea that some rereplication is occurring. The loss of viability in this experiment was smaller than the loss of viability seen in Figure 6 with strain YSH211 [MET-CDC6-NLS ORC2* ORC6* MCM7-nls(3A)], but the experiment of Figure 6 was done with arrested cells rather than asynchronous cells as here.
Table 4.
Loss of viability of ORC2* ORC6* and CDC6* cells over-expressing replication proteins
| Strain | Replication proteins | Cell count (×107/ml) |
No. of colonies |
||
|---|---|---|---|---|---|
| 0 h | 4 h | 0 h | 4 h | ||
| ORC2* ORC6*(St. 307) | GAL-DPB11 | 1.7 | 1.8 | 506 | 495 |
| GAL-SLD2 | 1.8 | 1.9 | 717 | 680 | |
| GAL-PRI1 | 2.0 | 2.2 | 765 | 881 | |
| CDC6* (St. 145) | GAL-DPB11 | 2.6 | 2.7 | 950 | 870 |
| GAL-SLD2 | 3.5 | 3.6 | 1225 | 1164 | |
| GAL-PRI1 | 3.2 | 3.6 | 1080 | 1230 | |
| WT (St. 48) | GAL-DPB11 | 2.1 | 2.6 | 888 | 890 |
| GAL-SLD2 | 3.4 | 4.0 | 1330 | 1540 | |
| GAL-PRI1 | 3.3 | 3.6 | 1389 | 1403 | |
Log phase cells growing on sucrose were washed and resuspended in medium containing galactose (2%) and incubated for 4 h at 30°C. The number of cells at 0 h (the time of addition of galactose) and 4 h was counted using a Coulter Counter. Equal volumes were taken from 0- and 4-h cultures, diluted, and spread on plates containing glucose (2%). Number of colonies was counted after 3 d at 30°C. GAL-PRI1 is a control; it does not cause toxicity in any of these strains.
We also characterized the GAL overexpression strains using flow cytometry. All the strains showing overexpression induced lethality also showed a small shift in their flow cytometry profiles toward higher DNA contents (Figure 9 and data not shown), suggesting that they may be undergoing rereplication.
Figure 9.
Overexpression of DPB11 increases DNA content in ORC* and CDC6* strains. Strains YSH287, YSH268, YSH310, and YSH209 were grown to log phase in sucrose medium. Cells were washed then divided into two halves. One-half of the cells were incubated in the presence of sucrose (2%) and other half in galactose (2%) to induce replication genes for a further 4 h at 30°C. CDC6* expression from the MET promoter was induced in YSH268 and YSH209 by growing the cells in medium lacking methionine. After 4 h, cells were fixed and stained with propidium iodide, and DNA contents were measured by flow cytometry. The percentage of cells containing more than 2C DNA was calculated using CellQuest software (Becton-Dickinson, Lincoln Park, NJ). Each panel contains, first, an experimental trace and then the control (uninduced) trace for the same strain.
To further characterize this possible rereplication, we transformed some of the GAL overexpression strains with DDC2-GFP. DDC2 is involved in a DNA damage checkpoint and in DNA repair (Melo et al., 2001). The Ddc2 protein is recruited to sites of DNA damage, including damage caused by rereplication (Archambault et al., 2005). Thus, rereplication (and other kinds of DNA damage) causes formation of foci of Ddc2. Some Ddc2 foci are seen even in cultures of normal cells, because DNA damage sometimes occurs during a normal S-phase. However, both the frequency of foci, and the number of foci per cell, increase with increased DNA damage.
Indeed, we found that both the frequency of foci, and the number of foci per cell, were higher in the strains that had overexpression induced lethality (Table 5). In fact, there was a perfect correlation between overexpression induced lethality, increased DNA content, and increased Ddc2 foci (Table 5 and data not shown). This suggests that these strains are suffering DNA damage, and this is consistent with the idea that they may be rereplicating to some degree.
DISCUSSION
CDK activity works by multiple mechanisms to inhibit association of Cdc6 with chromatin. CDK activity regulates transcription, degradation, and nuclear localization of Cdc6 (Bueno and Russell, 1992; Zwerschke et al., 1994; Piatti et al., 1995; Jong et al., 1996; Drury et al., 1997, 2000; Elsasser et al., 1999). In addition, at least in vitro, phosphorylation allows Cdc6 to bind to Clb2-Cdc28, and this prevents binding to ARS1 DNA beads (Mimura et al., 2004). Here, we find that even when forced into the nucleus, wild-type Cdc6 will not associate with chromatin, whereas Cdc6*, lacking CDK phosphorylation sites, will associate efficiently with chromatin. This suggests that the CDK phosphorylation sites of Cdc6 are rather directly involved in controlling assembly of the pre-RC. One possibility is that the phosphates sterically hinder an interaction between Cdc6 and some other protein at the origin. A second possibility is that phosphorylated Cdc6 is sequestered by Clb2-Cdc28 (Mimura et al., 2004). A third possibility is that Cdc6 (but not Cdc6*) brings Clb2-Cdc28 to the origin, which then phosphorylates still other proteins, lowering the affinity of the origin complex for Cdc6 (see below). Cdc6* can be reloaded onto G2/M chromatin very efficiently, and yet no detectable rereplication occurs in an otherwise wild-type strain, showing that Cdc6* reloading alone is not sufficient for rereplication.
Wild-type Cdc6 will also reload onto chromatin during G2/M (Tanaka et al., 1997) and cause rereplication in an ORC2* ORC6* strain (Figures 5 and 6) if, but only if, it is overexpressed. We believe this is because Cdc6 is a CDK inhibitor (Bueno and Russell, 1992; Calzada et al., 2001). When overexpressed, it inhibits CDK, and so may allow the accumulation of some unphosphorylated Cdc6. Presumably, this unphosphorylated Cdc6 can then reload onto chromatin just as if it were Cdc6*. In our studies, wild-type Cdc6 was only able to reload when Cdc6 expression was sufficiently high to cause an abnormal bud morphology suggestive of inhibition of CDK activity (Nugroho and Mendenhall, 1994; Elsasser et al., 1996; Verma et al., 1997). Expression of Cdc6* had relatively little effect on bud morphology, presumably because Cdc6* does not bind Cdc28 very well (Wolf et al., 1999; Mimura et al., 2004; Supplementary Figure 10) and so cannot inhibit it. Cdc6* reloaded onto chromatin under conditions where bud morphology was normal.
The fission yeast Schizosaccharomyces pombe is more permissive for rereplication than S. cerevisiae. Overexpression of either Cdc18 (the homolog of Cdc6) or of the CDK inhibitor Rum1 (the homolog/analog of Sic1) is sufficient for rereplication in S. pombe (Kelly et al., 1993; Moreno and Nurse, 1994; Nishitani and Nurse, 1995; Lopez-Girona et al., 1998), whereas the equivalent manipulations do not cause rereplication in S. cerevisiae. It is still somewhat unclear to what extent the rereplication caused by overexpression of Cdc18 is due to its activity as a CDK inhibitor, versus its activity as a replication inititator.
After Cdc6 has been recruited to origin and loaded Mcms, the Cdc6 falls off the chromatin before origins fire (Figure 4). During this period between α-factor release and S, there is little or no Cdc6 or Cdc6* on the chromatin, even in the MET-CDC6* strain constitutively expressing nuclear Cdc6* (Figure 4). We do not understand this G1-phase low-Cdc6 window. There is some decrease in the amount of Cdc6 or Cdc6* in whole cell extracts during this time, but this decrease does not seem sufficient to fully explain the larger decrease in Cdc6 associated with chromatin, so there may be some novel block to reloading Cdc6 before S-phase. One possibility is that once Mcms have been assembled onto the origin, the Mcms themselves (or a larger complex dependent on the Mcms) block the binding of Cdc6, and the site for Cdc6 binding is only revealed again after firing, when the Mcms have moved away from the origin.
Previous investigators have shown that rereplication is promoted by phosphorylation site mutants of Cdc6 (Nguyen et al., 2001; Vas et al., 2001; Mimura et al., 2004; Wilmes et al., 2004), and interactions have been seen between alleles of ORC6 and CDC6 (Wilmes et al., 2004), but results differ in detail. Nguyen et al. and Wilmes et al. found that an ectopic NLS on Mcm7 was also required for rereplication, whereas we and Mimura et al. saw no effect of Mcm7-NLS. However, the experiments differed in at least two ways. First, Nguyen et al. and Wilmes et al. used mutants of Cdc6 lacking their N-termini and hence lacking the natural nuclear localization signal. In contrast, our alleles of CDC6 possessed their natural NLS. (Mimura et al. used both kinds of Cdc6 mutants.). Furthermore our alleles of CDC6 contained an additional heterologous NLS. Possibly Cdc6 and Mcms enter the nucleus as a complex, and an NLS on either Cdc6 or Mcm might suffice for nuclear localization of both.
Second, Nguyen et al. (2001) and Wilmes et al. (2004) found dependence of rereplication on Mcm7-NLS using flow cytometry to monitor DNA content (a relatively insensitive assay), whereas we and Mimura et al. (2004) saw the lack of dependence on Mcm7-NLS in assays that measured cell viability (a probably more sensitive assay). Thus it may be that rereplication does not absolutely require Mcm7-NLS, but is more extensive (and therefore easier to see by flow cytometry) in its presence.
Rereplication is extremely toxic. Almost all cells induced to rereplicate die, even if the CDC6 inducing rereplication is turned off quickly after induction. Green and Li (2005) have shown that rereplication leads to double-strand DNA breaks, presumably the cause of the lethality.
One of our findings is that the CDC6* mutation is strongly synergistic with the ORC2* ORC6* mutations. Either alone is quite healthy with little if any phenotype, whereas the combination is completely lethal, even though Cdc6* is being expressed at extremely low levels (undetectable by Western), and even though a completely wild-type allele of CDC6 is present in the strain. An obvious kind of model is that Cdc6* allows the illegitimate binding of one set of proteins to the origin, whereas Orc2* Orc6* allows the binding of a different set of proteins, and together these two sets of proteins allow rereplication. We tested this idea by overexpressing various replication proteins to see if we could induce rereplication. Indeed, overexpression of several important initiation proteins such as Dpb11 and Sld2 were toxic, and the Ddc2 repair foci and increased DNA contents in these strains suggested that rereplication may have occurred. However, the initiation proteins that were most toxic in the CDC6* strain were also the most toxic in the ORC2* ORC6* strains, and this suggests that CDC6* and ORC2* ORC6* are acting in similar yet redundant ways. Interestingly, the two initiator proteins with the strongest phenotypes in our assays, Dpb11 and Sld2, are known to bind each other, and are intimately involved in the initiation of replication (Kamimura et al., 1998; Masumoto et al., 2000, 2002). Sld2 is likely one of the key initiators activated by CDK (Masumoto et al., 2002).
As an admittedly speculative model to explain how CDC6* could be redundant with ORC2* ORC6*, we propose the following: Rereplication is normally prevented because the cell localizes Clb5-Cdc28 to the origin via Orc6 (Wilmes et al., 2004), and redundantly (we propose), the cell localizes Clb2-Cdc28 (and Clb1-Cdc28) to the origin via Cdc6. Mimura et al. (2004) showed binding between Cdc6 and Clb2, and we suggest that this binding might allow Cdc6 to bring Clb2-Cdc28 to the origin. Once at the origin, both Clb5-Cdc28 and Clb2-Cdc28 phosphorylate a partially overlapping set of initiator proteins, preventing rereplication. Phosphorylation site mutants of Cdc6 (which fail to bind Clb2) are synthetically lethal with clb5 deletion mutants (Wilmes et al., 2004) because there is then no way to localize any Clb-CDK activity to the origin. Similarly, CDC6* is synthetically lethal with ORC2* ORC6* because Cdc6* cannot target Clb2 to the origin, and Orc2 and Orc6 are the most important targets of origin-localized Clb5-Cdc28 (and furthermore may aid in the binding of Clb5 to Orc). Because CDC6* mutants and ORC2* ORC6* mutants have essentially the same molecular defect (i.e., decreased phosphorylation of origin proteins), they are sensitive to overexpression of essentially the same initiator proteins. Their sensitivities are not exactly the same, because the phosphorylation events they lack are not exactly the same.
There are at least two objections to this model. First, a clb2 clb5 double mutant is viable (Epstein and Cross, 1992). But Clb1 may be able to replace Clb2 for preventing rereplication.
Second, Mimura et al. (2004) have suggested that Clb2 sequesters Cdc6 and prevents it from binding at the origin, the opposite of our proposal. But mechanistically, it is unclear how this proposed sequestration could work. Clb2 binds to the extreme N-terminus of Cdc6, but this N-terminal region is not needed for Cdc6 to bind to the ORC complex, so there is no obvious reason why a Cdc6-Clb2 complex should fail to bind ORC. Thus we offer a reinterpretation of the results of Mimura et al. Perhaps Cdc6 at the origin does bind Clb2-Cdc28, and at least temporarily, does bring Clb2-Cdc28 to the (hypophosphorylated) ORC complex. The Clb2-Cdc28 then phosphorylates various proteins, and this phosphorylated origin complex is now inhospitable toward Cdc6 binding, and the Cdc6-Clb2-Cdc28 complexes falls off. So, at equilibrium, the net effect is that the Cdc6-Clb2-Cdc28 complex is not bound near origins, and it is this equilibrium situation that Mimura et al. observed in their experiments; i.e., at equilibrium, Clb2 does indeed prevent Cdc6 from binding at origins. Nevertheless, our model suggests that there was an intermediate period when Cdc6 and Clb2-Cdc28 were at the origin, and during this period, origin proteins important for preventing rereplication were phosphorylated by Clb2-Cdc28. One prediction of this model is that if Clb2 were artificially tethered to the origin, it would prevent rereplication in a CDC6* ORC2* ORC6* strain. A second more speculative prediction is that a CDC6* clb5 clb6 strain might be incapable of initiating replication, as Cdc28 might have no means of localizing to the origin.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gavin Sherlock for providing CDC6*, for preliminary experiments, and for insightful discussions. We thank Padmini Arora for help with chromatin association experiments. We thank Joachim Li for providing key strains and plasmids and for insightful discussions. We thank B. Stillman for anti-Orc3 and anti-Mcm antibodies. We thank D. Toczyski for DDC2-GFP. We thank A. Rosebrock and F. Ferrezuelo for help with microarrays. This work was supported by National Institutes of Health Grant RO1 GM039978 to B.F.
Abbreviations used:
- CDK
cyclin dependent kinase
- ORC
origin recognition complex
- RC
replication complex.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-06-0544) on January 31, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Aparicio O. M., Weinstein D. M., Bell S. P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Archambault V., Ikui A. E., Drapkin B. J., Cross F. R. Disruption of mechanisms that prevent rereplication triggers a DNA damage response. Mol. Cell. Biol. 2005;25:6707–6721. doi: 10.1128/MCB.25.15.6707-6721.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. P., Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Bueno A., Russell P. Dual functions of CDC6, a yeast protein required for DNA replication also inhibits nuclear division. EMBO J. 1992;11:2167–2176. doi: 10.1002/j.1460-2075.1992.tb05276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzada A., Sacristan M., Sanchez E., Bueno A. Cdc6 cooperates with Sic1 and Hct1 to inactivate mitotic cyclin-dependent kinases. Nature. 2001;412:355–358. doi: 10.1038/35085610. [DOI] [PubMed] [Google Scholar]
- Cocker J. H., Piatti S., Santocanale C., Nasmyth K., Diffley J. F. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- Detweiler C. S., Li J. J. Cdc6p establishes and maintains a state of replication competence during G1 phase. J. Cell Sci. 1997;110(Pt 6):753–763. doi: 10.1242/jcs.110.6.753. [DOI] [PubMed] [Google Scholar]
- Diffley J. F. Once and only once upon a time: specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- Donovan S., Harwood J., Drury L. S., Diffley J. F. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc. Natl. Acad. Sci. USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury L. S., Perkins G., Diffley J. F. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury L. S., Perkins G., Diffley J. F. The cyclin-dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr. Biol. 2000;10:231–240. doi: 10.1016/s0960-9822(00)00355-9. [DOI] [PubMed] [Google Scholar]
- Edgington N. P., Futcher B. Relationship between the function and the location of G1 cyclins in S. cerevisiae. J. Cell Sci. 2001;114:4599–4611. doi: 10.1242/jcs.114.24.4599. [DOI] [PubMed] [Google Scholar]
- Elsasser S., Chi Y., Yang P., Campbell J. L. Phosphorylation controls timing of Cdc6p destruction: a biochemical analysis. Mol. Biol. Cell. 1999;10:3263–3277. doi: 10.1091/mbc.10.10.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S., Lou F., Wang B., Campbell J. L., Jong A. Interaction between yeast Cdc6 protein and B-type cyclin/Cdc28 kinases. Mol. Biol. Cell. 1996;7:1723–1735. doi: 10.1091/mbc.7.11.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein C. B., Cross F. R. CLB5, a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- Green B. M., Li J. J. Loss of rereplication control in Saccharomyces cerevisiae results in extensive DNA damage. Mol. Biol. Cell. 2005;16:421–432. doi: 10.1091/mbc.E04-09-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green B. M., Morreale R. J., Ozaydin B., Derisi J. L., Li J. J. Genome-wide mapping of DNA synthesis in Saccharomyces cerevisiae reveals that mechanisms preventing reinitiation of DNA replication are not redundant. Mol. Biol. Cell. 2006;17:2401–2414. doi: 10.1091/mbc.E05-11-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey S., Schneider B. L., Schieltz D. M., Yates J. R., Futcher B. A novel multiple affinity purification tag and its use in identification of proteins associated with a cyclin-CDK complex. Nucleic Acids Res. 2001;29:E24. doi: 10.1093/nar/29.4.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jong A., Young M., Chen G. C., Zhang S. Q., Chan C. Intracellular location of the Saccharomyces cerevisiae CDC6 gene product. DNA Cell Biol. 1996;15:883–895. doi: 10.1089/dna.1996.15.883. [DOI] [PubMed] [Google Scholar]
- Kamimura Y., Masumoto H., Sugino A., Araki H. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol. Cell. Biol. 1998;18:6102–6109. doi: 10.1128/mcb.18.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T. J., Brown G. W. Regulation of chromosome replication. Annu. Rev. Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- Kelly T. J., Martin G. S., Forsburg S. L., Stephen R. J., Russo A., Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Liang C., Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Weinreich M., Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona A., Mondesert O., Leatherwood J., Russell P. Negative regulation of Cdc18 DNA replication protein by Cdc2. Mol. Biol. Cell. 1998;9:63–73. doi: 10.1091/mbc.9.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K. Q., Elsasser S., Chang D. C., Campbell J. L. Regulation of the localization and stability of Cdc6 in living yeast cells. Biochem. Biophys. Res. Commun. 2003;306:851–859. doi: 10.1016/s0006-291x(03)01082-9. [DOI] [PubMed] [Google Scholar]
- Masumoto H., Muramatsu S., Kamimura Y., Araki H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 2002;415:651–655. doi: 10.1038/nature713. [DOI] [PubMed] [Google Scholar]
- Masumoto H., Sugino A., Araki H. Dpb11 controls the association between DNA polymerases alpha and epsilon and the autonomously replicating sequence region of budding yeast. Mol. Cell. Biol. 2000;20:2809–2817. doi: 10.1128/mcb.20.8.2809-2817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo J. A., Cohen J., Toczyski D. P. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura S., Seki T., Tanaka S., Diffley J. F. Phosphorylation-dependent binding of mitotic cyclins to Cdc6 contributes to DNA replication control. Nature. 2004;431:1118–1123. doi: 10.1038/nature03024. [DOI] [PubMed] [Google Scholar]
- Moreno S., Nurse P. Regulation of progression through the G1 phase of the cell cycle by the rum1+ gene. Nature. 1994;367:236–242. doi: 10.1038/367236a0. [DOI] [PubMed] [Google Scholar]
- Nguyen V. Q., Co C., Li J. J. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- Nishitani H., Nurse P. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell. 1995;83:397–405. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- Nugroho T. T., Mendenhall M. D. An inhibitor of yeast cyclin-dependent protein kinase plays an important role in ensuring the genomic integrity of daughter cells. Mol. Cell. Biol. 1994;14:3320–3328. doi: 10.1128/mcb.14.5.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva A., Rosebrock A., Ferrezuelo F., Pyne S., Chen H., Skiena S., Futcher B., Leatherwood J. The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol. 2005;3:e225. doi: 10.1371/journal.pbio.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatti S., Bohm T., Cocker J. H., Diffley J. F., Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- Piatti S., Lengauer C., Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional' anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez M., Calzada A., Bueno A. The Cdc6 protein is ubiquitinated in vivo for proteolysis in Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:9092–9097. doi: 10.1074/jbc.274.13.9092. [DOI] [PubMed] [Google Scholar]
- Santocanale C., Diffley J. F. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J. 1996;15:6671–6679. [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Knapp D., Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Tanny R. E., MacAlpine D. M., Blitzblau H. G., Bell S. P. Genome-wide analysis of re-replication reveals inhibitory controls that target multiple stages of replication initiation. Mol. Biol. Cell. 2006;17:2415–2423. doi: 10.1091/mbc.E05-11-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Vas A., Mok W., Leatherwood J. Control of DNA rereplication via Cdc2 phosphorylation sites in the origin recognition complex. Mol. Cell Biol. 2001;21:5767–5777. doi: 10.1128/MCB.21.17.5767-5777.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R., Feldman R. M., Deshaies R. J. SIC1 is ubiquitinated in vitro by a pathway that requires CDC4, CDC34, and cyclin/CDK activities. Mol. Biol. Cell. 1997;8:1427–1437. doi: 10.1091/mbc.8.8.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M., Liang C., Stillman B. The Cdc6p nucleotide-binding motif is required for loading mcm proteins onto chromatin. Proc. Natl. Acad. Sci. USA. 1999;96:441–446. doi: 10.1073/pnas.96.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes G. M., Archambault V., Austin R. J., Jacobson M. D., Bell S. P., Cross F. R. Interaction of the S-phase cyclin Clb5 with an “RXL” docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 2004;18:981–991. doi: 10.1101/gad.1202304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D. A., McKeon F., Jackson P. K. Budding yeast Cdc6p induces re-replication in fission yeast by inhibition of SCF(Pop)-mediated proteolysis. Mol. Gen. Genet. 1999;262:473–480. doi: 10.1007/s004380051108. [DOI] [PubMed] [Google Scholar]
- Zhu G., Spellman P. T., Volpe T., Brown P. O., Botstein D., Davis T. N., Futcher B. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature. 2000;406:90–94. doi: 10.1038/35017581. [DOI] [PubMed] [Google Scholar]
- Zwerschke W., Rottjakob H. W., Kuntzel H. The Saccharomyces cerevisiae CDC6 gene is transcribed at late mitosis and encodes a ATP/GTPase controlling S phase initiation. J. Biol. Chem. 1994;269:23351–23356. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.