Abstract
Using phage display, we identified Na+/H+ exchanger regulatory factor (NHERF)-2 as a novel binding partner for the cadherin-associated protein, β-catenin. We showed that the second of two PSD-95/Dlg/ZO-1 (PDZ) domains of NHERF interacts with a PDZ-binding motif at the very carboxy terminus of β-catenin. N-cadherin expression has been shown to induce motility in a number of cell types. The first PDZ domain of NHERF is known to bind platelet-derived growth factor-receptor β (PDGF-Rβ), and the interaction of PDGF-Rβ with NHERF leads to enhanced cell spreading and motility. Here we show that β-catenin and N-cadherin are in a complex with NHERF and PDGF-Rβ at membrane ruffles in the highly invasive fibrosarcoma cell line HT1080. Using a stable short hairpin RNA system, we showed that HT1080 cells knocked down for either N-cadherin or NHERF had impaired ability to migrate into the wounded area in a scratch assay, similar to cells treated with a PDGF-R kinase inhibitor. Cells expressing a mutant NHERF that is unable to associate with β-catenin had increased stress fibers, reduced lamellipodia, and impaired cell migration. Using HeLa cells, which express little to no PDGF-R, we introduced PDGF-Rβ and showed that it coimmunoprecipitates with N-cadherin and that PDGF-dependent cell migration was reduced in these cells when we knocked-down expression of N-cadherin or NHERF. These studies implicate N-cadherin and β-catenin in cell migration via PDGF-R–mediated signaling through the scaffolding molecule NHERF.
INTRODUCTION
β-Catenin is a member of the armadillo family of proteins, which contain central repeat elements known as armadillo repeats. β-Catenin functions in the adherens junction where it creates a link between cadherins and the actin cytoskeleton through its interactions with α-catenin. β-Catenin also functions in Wnt signaling and contains a transcriptional activation domain in its carboxy-terminal region (Brembeck et al., 2006). Structurally, β-catenin consists of a central armadillo repeat domain flanked by unique amino- and carboxy-terminal domains. The armadillo repeat domain of β-catenin mediates interactions with a number of well-characterized partners, including cadherins, adenomatous polyposis coli (APC), and T-cell factor (TCF) family members (Bienz, 2005). The amino-terminal domain of β-catenin interacts with α-catenin and contains amino acid residues important for regulating protein stability (Gottardi and Gumbiner, 2001). The current study was designed to further understand protein–protein interactions involving the carboxy terminus of β-catenin.
Using phage display we found that NHERF-2 (Na+/H+ exchanger regulatory factor 2) is a binding partner for the C-terminus of β-catenin. NHERF-2 contains two N-terminal PSD-95/Dlg/ZO-1 (PDZ) domains and a C-terminal region that binds to members of the ezrin/radixin/moesin (ERM) family of actin-associated proteins (Reczek and Bretscher, 1998; Yun et al., 1998). NHERF-2 is a scaffolding protein involved in cAMP-mediated regulation of Na+/H+ exchanger 3 (NHE3) (Lamprecht et al., 1998). NHE3 binds to the second PDZ domain of NHERF-2, creating a link between ezrin and NHE3 (Yun et al., 1998). In addition, NHERF-2 is involved in the regulation of proteins at the apical surface of epithelial cell membranes, such as ion transporters and transmembrane receptors (Shenolikar and Weinman, 2001; Voltz et al., 2001; Weinman et al., 2006).
NHERF-2 is 52% identical to another protein called NHERF-1. Structural studies of the interaction between NHERF-1 and the cystic fibrosis transmembrane conductance regulator (CFTR) revealed the specificity of NHERF PDZ domains for their partners (Karthikeyan et al., 2001). Both NHERF-1 and NHERF-2 bind to the very C-terminal amino acids (DTRL-COOH) of CFTR (Wang et al., 1998; Sun et al., 2000). PDZ domains typically bind to short sequences, often at the C terminus of their partner, that can be grouped into classes of consensus motifs based on binding specificity (Songyang et al., 1997). The C-terminal amino acids of CFTR belong to the class I PDZ-binding consensus, which is characterized by the sequence (S/T)X(V/I/L), where X is any amino acid. The C-terminal sequence of β-catenin is DTDL, which makes it a potential binding partner for class I PDZ domains. Here, we show that β-catenin and NHERF-2 interact at the plasma membrane of epithelial cells and that this association requires the second PDZ domain of NHERF-2 and the C-terminal PDZ-binding motif of β-catenin.
The platelet derived growth factor receptor (PDGF-R) is a dimeric receptor tyrosine kinase made up of α and/or β isoforms. Signaling downstream of this receptor in response to binding of the ligand PDGF leads to cell growth, actin reorganization, cell migration, and differentiation (Heldin et al., 1998). Both PDGF-Rα and β isoforms contain the C-terminal PDZ-binding motif DSFL. PDGF-Rβ has been shown to interact with the first PDZ domain of NHERF-1 and NHERF-2 via its C terminus, and this interaction has been shown to regulate PDGF-R signaling and actin cytoskeletal reorganization (Maudsley et al., 2000; Demoulin et al., 2003; James et al., 2004).
Cadherins form a family of cell surface, transmembrane proteins that mediate cell–cell recognition and adhesion. E-cadherin typically is expressed by epithelial cells, whereas N-cadherin is found in multiple cell types, including nerve, myocardial, and mesenchymal cells. In vitro, human tumor cell lines misexpressing N-cadherin exhibit increased migration and invasion (Islam et al., 1996; Nieman et al., 1999; Hazan et al., 2000). The influence of N-cadherin on stimulating cell migration and invasion is thought to involve its interaction with another receptor tyrosine kinase, fibroblast growth factor receptor (FGFR) (Nieman et al., 1999; Suyama et al., 2002). HT1080 is a highly motile and invasive fibrosarcoma cell line that expresses PDGF-R and N-cadherin. Here, we show that NHERF-2 links the N-cadherin–β-catenin complex to PDGF-Rβ in HT1080 cells at lamellipodia and that this complex contributes to actin cytoskeletal changes and cell migration.
MATERIALS AND METHODS
Cell Culture
A431 cells (ATCC CRL-1555; American Type Culture Collection, Manassas, VA), HT1080 cells (ATCC CRL-121), and the human colon carcinoma cell line SW707 cells (Leibovitz et al., 1976) were grown in DMEM (Sigma-Aldrich, St. Louis, MO) with 10% fetal bovine serum (FBS; Hyclone Laboratories, Logan, UT). Batt cells (Fleury-Feith et al., 1995) were a kind gift from Dr. Marie-Claude Jaurand at Institut National de la Santé et de la Recherche Médicale-Equipe Mixte Inserm 99.09 and were grown in RPMI 1640 medium (Sigma-Aldrich) with 10% FBS. The mouse myeloma NS1 (ATCC TIB-18) and the hybridoma cell lines were grown in HY medium (Sigma-Aldrich) with 20% FBS. HeLa cells were grown in minimal essential medium (Sigma-Aldrich) with 10% FBS. Human microvascular endothelial cell (HMEC)-1, a simian virus 40 immortalized human dermal microvascular endothelial cell line, was obtained from The Center for Disease Center, Atlanta, GA, and grown in MCDB-131 medium (Sigma-Aldrich) containing 1 μg/ml hydrocortisone and 10 ng/ml epidermal growth factor supplemented with 10% fetal bovine serum (Ades et al., 1992).
Antibodies
Mouse monoclonal anti-β-catenin (15B8), anti-E-cadherin (4A2), anti-N-cadherin (13A9), and anti-α-catenin (1G5) have been described previously (Johnson et al., 1993; Sacco et al., 1995). Mouse monoclonal antibody (mAb) against the myc tag (9E10) was a kind gift of Dr. Kathy Green (Northwestern University, Chicago, IL). Mouse monoclonal anti-glutathione S-transferase (GST) antibody was purchased from Zymed Laboratories (South San Francisco, CA). Mouse monoclonal anti-ezrin was purchased from BD Transduction Laboratories (San Diego, CA). Rabbit anti-c-myc was purchased from Upstate Biotechnology (Charlottesville, VA). Rabbit anti-β-catenin was purchased from Sigma-Aldrich. Mouse anti-NHERF-1 was purchased from Abcam (Cambridge, MA). Mouse monoclonal antibodies against NHERF-2 were generated as described previously (Wahl, 2002), using full-length NHERF-2 fused to maltose binding protein (MBP). Antibodies were mapped to the region of the first PDZ domain (33E2) and the second PDZ domain (32B6). Anti-β-catenin beads were created by conjugating 15B8 to Sepharose using the protocol provided with CNBr-activated Sepharose 4B (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). A rabbit polyclonal PDGF-Rβ antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA) (sc-339). Mouse monoclonal anti-birch profilin tag (4A6) was a kind gift of Dr. Manfred Rudiger (Technical University of Braunschweig, Braunschweig, Germany).
DNA Constructs and Transfections
A restriction fragment encoding AA 695-781 of chicken β-catenin was 6XHIS tagged by subcloning into pHIS parallel-1 (Sheffield et al., 1999). NHERF-1 and NHERF-2 clones were obtained from American Type Culture Collection (BE250116 and BE301870, respectively). Full-length NHERF-1 (AA1-358) and full-length NHERF-2 (AA1-337) as well as deletion constructs of NHERF-2 encoding the indicated amino acids, PDZ I (1-95), PDZII (143-247), PDZII+C-term (143-337), PDZI+II (8-247), and C-term (248-337) were subcloned in frame in the pGST parallel vector (Sheffield et al., 1999). Full-length NHERF-2 as well as deletion constructs encoding the indicated amino acids, PDZ I (1-95), PDZII (143-247), PDZI+II (1-247), PDZII + 1/2C-term (143-280), PDZII+C-term (143-337), and C terminus (248–337), were N-terminally 2Xmyc tagged in pSPUTK (Falcone and Andrews, 1991) and subsequently subcloned into the mammalian expression vector pLKpac (Hirt et al., 1992; Islam et al., 1996). All constructs were created using convenient restriction sites, except PDZII, PDZII + 1/2C-term, and PDZI+II, which were synthesized by polymerase chain reaction (PCR) before adding the 2Xmyc tag. Full-length β-catenin was N-terminally 2Xmyc tagged and subcloned into pLKpac. Three β-catenin mutant constructs containing dialanine mutations in the C terminus (W776A/F777A, D778A/T779A, and D780A/L781A) were created using PCR and subcloned into pLKpac (details available upon request). Solutions from the Mammalian Transfection kit (Stratagene, La Jolla, CA) were used to stably transfect A431 and Batt cells. Cells were grown to ∼20% confluence, transfected with 10 μg of DNA, and selected in 2 μg/ml puromycin.
Myc-tagged full-length NHERF-2 was subcloned into the viral expression vector LZRS-MS-neo (Ireton et al., 2002), transfected into Phoenix 293 cells (Grignani et al., 1998), and supernatant was collected for infection of HT1080 cells as described previously (Maeda et al., 2005). A cDNA clone encoding human PDGF-Rβ was obtained from Open Biosystems (Huntsville, AL) (MHS1010). The clone contained a mutation (S180F) and was corrected using the QuikChange site-directed mutagenesis kit (Stratagene). Wild-type PDGF-Rβ was subcloned into a derivative of LZRS-MS-neo that confers resistance to hygromycin. Sequence coding for the last 53 amino acids of PDGF-Rβ was amplified by PCR and subcloned into a pMBP parallel vector (Sheffield et al., 1999) tagging the construct with MBP. The C-terminal mutant form of β-catenin D778A/T779A was tagged with a birch profilin epitope (Rudiger et al., 1997) and subcloned into a LZRS-MS-neo derivative that confers resistance to puromycin (details available upon request). N-cadherin (Maeda et al., 2005) and NHERF-1 expression were knocked down using the retroviral shRNA expression system pSuperRetro (OligoEngine, Seattle, WA). The shRNA sequence for N-cadherin has been published previously (Maeda et al., 2005), and the sequence of NHERF-1 targeted by shRNA was CTGACGAGTTCTTCAAGAA.
Phage Display Analysis
The 6XHIS-tagged carboxy terminus of β-catenin was expressed in JM109 DE3 cells (Promega, Madison, WI). Bacteria were lysed with lysis buffer (5 mM Na2HPO4, pH 7.2, 0.875 g/l NaCl, 0.125% Tween 20, and 0.035% β-mercaptoethanol), and bait protein was immobilized on Reacti-Bind metal chelate plates (Pierce Chemical, Rockford, IL), and the plate was washed with Tris-buffered saline/Tween 20 (TBST) (10 mM Tris-HCl, pH 8.0, 9 g/l NaCl, and 0.05% Tween 20). The T7Select system was used to screen a human breast cDNA T7 phage display library, both purchased from Novagen (Madison, WI). Four rounds of biopanning were done by washing bound phage with TBST and eluting with 3 M urea. PCR amplification was done on positive phage plaque inserts by using the T7SelectUP and T7SelectDOWN primer set, and the products were analyzed by sequencing.
In Vitro Binding Assays
GST-tagged proteins consisting of NHERF-1 and various regions of NHERF-2, including full-length protein, the first PDZ domain, the second PDZ domain including the C terminus, both PDZ domains alone, and the C terminus alone, were purified on a glutathione column (Sigma-Aldrich), washed with Tris-buffered saline (10 mM Tris-HCl, pH 8.0, and 9 g/l NaCl) and eluted with 10 mM reduced glutathione in 50 mM Tris-HCl, pH 8.0. Equal amounts of purified protein were incubated with glutathione resin, washed with deoxycholate (DOC) (50 mM Tris-HCl, pH 7.5, 9 g/l NaCl, 0.1% Triton X-100, 1% deoxycholic acid, and 0.01% SDS), and incubated with SW707 cytosolic extract. Beads were again washed in DOC, boiled for 10 min in Laemmli sample buffer (Laemmli, 1970), resolved by SDS-PAGE, and transferred to nitrocellulose.
Immunoprecipitation and Western Blotting
All polypropylene tubes were rinsed with 0.01% Triton X-100 and dried before use in immunoprecipitations. To induce expression from pLK plasmids, dexamethasone was added (10−7 M) to culture media 24 h before extraction. Cell extracts for immunoprecipitation were made by washing the cells in phosphate-buffered saline and extracting in TNE (10 mM Tris acetate, pH 8.0, 0.5% NP-40, 1 mM EDTA, and 2 mM phenylmethylsulfonyl fluoride). Insoluble material was pelleted by centrifugation at 14,000 × g for 15 min at 4°C, and the supernatant was used for immunoprecipitation. For membrane fractionation, cells were suspended in TE (TNE without NP-40) and lysed by Dounce homogenization before centrifugation. The remaining pellet was washed once in phosphate-buffered saline (PBS) and resuspended in TNE to solubilize membrane proteins. Then, 300 μl of hybridoma-conditioned medium was added to 50 μl of packed anti-mouse IgG affinity gel (MP Biomedicals, Aurora, OH) and gently mixed at 4°C for 30 min. Excess antibody solution was removed, equal volumes of cell extract were added, and mixing continued for 30 min. For anti-myc immunoprecipitations (Figure 2C), the volume of lysate used was normalized for myc-tagged protein expression in A431 cells. Immune complexes were washed five times with TBST. After the final wash, the packed beads were resuspended in 50 μl of 2× Laemmli sample buffer, boiled for 5 min, and the proteins were resolved by SDS-PAGE. Proteins were electrophoretically transferred overnight to nitrocellulose membranes and immunoblotted as described previously (Wahl et al., 2003).
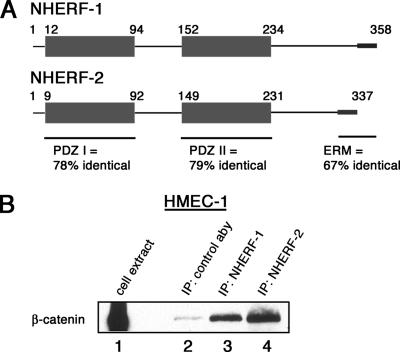
Figure 2.
NHERF-1 and NHERF-2 share sequence homology, and both associate with β-catenin. (A) Schematic diagram of NHERF-1 and NHERF-2. Highlighted are the conserved domains and sequence identities. (B) NHERF coimmunoprecipitates with β-catenin. NHERF-1 and NHERF-2 were immunoprecipitated from total HMEC-1 cell lysate. The immunoprecipitates were resolved by SDS-PAGE and immunoblotted for β-catenin. Lane 1 shows the amount of β-catenin in total cell lysate.
Immunofluorescence
Cells were grown on glass coverslips to 80% confluence and dexamethasone was added (10−7 M) as needed to the culture media 24 h before fixation in 3.7% formaldehyde for 15 min and processed as described previously (Wahl et al., 2003). Images were collected on a Zeiss Axiovert 200 M equipped with an ORCA-ER (Hamamatsu, Houston, TX) digital camera and processed using SlideBook software (Intelligent Imagining Innovations, Santa Monica, CA). Z-stacks of images were taken in 0.5-μm increments and digitally deconvolved with SlideBook software. Analysis of the percentage of cells with stress fibers was performed by counting the cells in random fields of view until the cell total reached 300 while scanning multiple planes of focus to count the number of cells with stress fibers. Cells with stress fibers throughout the cell, not just at the most basal plane of focus, where counted as having enhanced stress fibers (Supplemental Figure 2).
Migration Assays
To analyze cell migration, confluent monolayers of cells on tissue culture dishes were wounded by manually scratching with a pipette tip, washed with PBS, and incubated at 37°C in complete media. At the indicated times, phase-contrast images at specific wound sites were captured. The PDGF-R ligand, PDGF-BB (P4056), was from Sigma-Aldrich. The PDGF-R kinase inhibitor III (521232; Calbiochem, San Diego, CA) was added at 100 μM dimethyl sulfoxide. The number of pixels in the denuded area was calculated using Adobe Photoshop (Adobe Systems, San Jose, CA). Three independent experiments were averaged, and the SD of the final time point is represented with error bars.
RESULTS
NHERF-2 Binds to β-Catenin
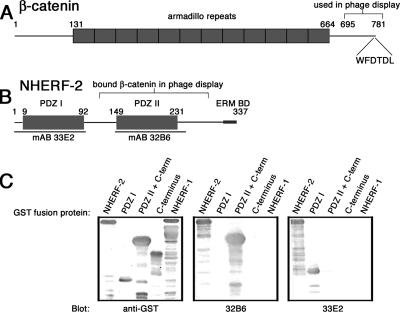
To explore novel functions for β-catenin, we used its carboxy-terminal 87 amino acids to screen a human phage display cDNA library. A clone encoding a portion of NHERF-2 that included the second of two PDZ domains and a portion of the C terminus was identified as a potential β-catenin binding partner. Figure 1 presents a diagram of β-catenin indicating the region used in biopanning (Figure 1A) and a diagram of NHERF-2 showing the portion identified by phage display (Figure 1B). The tax-interacting protein-1 (TIP-1) was also identified in this screen, giving us confidence in our screening method, because TIP-1 is a PDZ domain-containing protein shown by others to associate with the carboxy terminus of β-catenin (Kanamori et al., 2003).
Figure 1.
NHERF-2 is a potential binding partner for β-catenin. Schematic diagrams of β-catenin (A) and NHERF-2 (B). Highlighted are the portion of β-catenin used as bait for phage display and the portion of NHERF-2 that bound to β-catenin in phage display as well as the potential PDZ-binding motif (WFDTDL) in last six AA of β-catenin and the ERM-binding site at the C terminus of NHERF-2. (C) Purified GST fusion proteins including various NHERF-2 domains (or NHERF-1 as a negative control) were resolved by SDS-PAGE and immunoblotted with anti-GST antibodies, mAb 32B6, and mAb 33E2. Highlighted in B are the regions of NHERF-2 recognized by the two mAbs.
To aid in assessing the physiological relevance of interactions between β-catenin and NHERF-2, we generated monoclonal antibodies against human NHERF-2 by immunizing mice with full-length NHERF-2 as described previously (Wahl, 2002). Two hybridomas were selected and epitope mapped using truncated versions of NHERF-2 (Figure 1C). Antibody 33E2 recognized a portion of NHERF-2 in the first PDZ domain, whereas antibody 32B6 recognized a region in the second PDZ domain (Figure 1B). Neither of these antibodies showed cross-reactivity to the NHERF-2 homologue NHERF-1.
Interactions with β-Catenin Involve PDZ II and the C Terminus of NHERF
To examine potential interactions between β-catenin and NHERF proteins, we used the immortalized endothelial cell line HMEC-1, which expresses both NHERF-1 and NHERF-2. Immunoprecipitations were done with a mAb to NHERF-1 (Abcam) and our monoclonal NHERF-2 antibody 32B6. Using HMEC-1 cell extract, we found that β-catenin coimmunoprecipitated with NHERF-1 and NHERF-2 (Figure 2B). It is not surprising that an interaction partner for one of the NHERF proteins also binds the other, because they are highly homologous. Figure 2A shows a schematic diagram of both NHERF isoforms and the amino acid sequence identity shared in their conserved PDZ domains as well as the ERM-binding domain. Because we identified NHERF-2 in our phage display screen, we focused our attention on the potential interaction between this isoform and β-catenin.
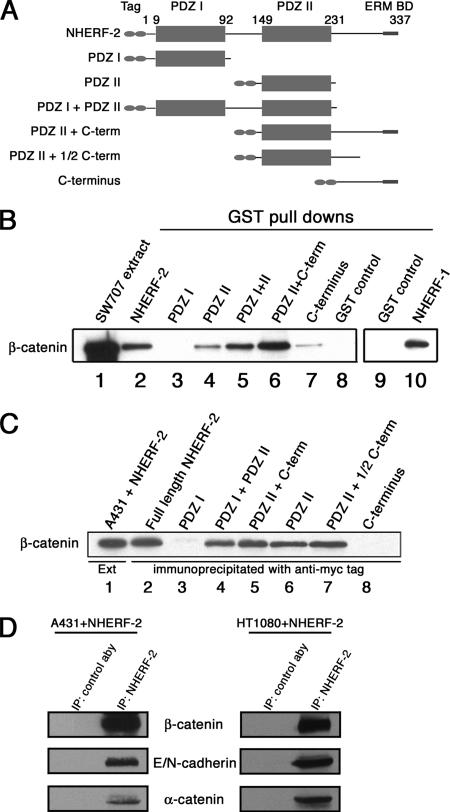
To characterize the interaction between β-catenin and NHERF-2, GST-tagged NHERF-2 constructs were immobilized on glutathione resin and incubated with SW707 cell extract. SW707 cells overexpress free cytosolic β-catenin, presumably due to a mutation in APC, and detergent-free extracts of these cells are a rich source of free β-catenin. The constructs used in this experiment are shown in Figure 3A, and the GST pull-downs are shown in Figure 3B. Lane 1 contains SW707 cell extract. Lanes 2–8 are pull-down assays by using GST fused to the various NHERF-2 constructs. β-Catenin bound effectively to full-length NHERF-1 (lane 10) and NHERF-2 as expected (lane 2), and it had no affinity for the first PDZ domain (lane 3). Constructs made up of PDZ II, PDZ I + PDZ II, or PDZ II + C terminus each efficiently pulled down β-catenin (lanes 4–6). The PDZ II domain containing the C-terminal portion of NHERF-2 was able to pull down the most β-catenin (lane 6). This is not surprising, because Yun et al. reported that the interaction of NHERF-2 with NHE3 requires both the PDZ II domain and the C terminus for highest affinity binding (Yun et al., 1998). In addition, the very C-terminal portion of NHERF-2 alone was able to pull down a small amount of β-catenin, suggesting there may be a weak β-catenin–binding domain present in this region (lane 7).
Figure 3.
NHERF-2 interacts with β-catenin in vitro and in vivo. (A) Schematic of tagged NHERF-2 proteins, indicating the tag, the two PDZ domains, and the ERM-binding domain (ERM BD). (B) An equal amount of each GST-tagged NHERF protein was immobilized on glutathione resin and incubated with SW707 cell lysate. The bound proteins were resolved by SDS-PAGE and immunoblotted for β-catenin (lanes 2–10). Lane 1 shows β-catenin in the SW707 extract. (C) A431 cells were transfected with 2× myc-tagged versions of the constructs shown in A. Cells were extracted, and lysate was normalized for myc-tagged protein expression, immunoprecipitated with anti-myc, resolved by SDS-PAGE, and immunoblotted for β-catenin. (D) Membrane fractions prepared from A431 cells or HT1080 cells expressing full-length myc-tagged NHERF-2 were extracted with NP-40, immunoprecipitated with anti-NHERF-1 (32B6) or control antibody, resolved by SDS-PAGE, and immunoblotted for β-catenin (15B8), E-cadherin (4A2), N-cadherin (13A9), and α-catenin (1G5).
To determine whether the interaction between β-catenin and NHERF-2 occurs in vivo, A431 cells, which do not express NHERF-2, were transfected with 2× myc-tagged versions of the constructs shown in Figure 3A. Extracts from transfected cells were immunoprecipitated with anti-myc and resolved by SDS-PAGE. Figure 3C shows that endogenous β-catenin coimmunoprecipitated with transfected full-length NHERF-2 (lane 2). In agreement with the GST pull-down experiments, β-catenin bound preferentially to the second PDZ domain of NHERF-2, because each of the proteins containing PDZ II was able to coimmunoprecipitate β-catenin (lanes 4–7), whereas the PDZ I domain alone did not bind to β-catenin (lane 3). In this in vivo experiment, the C terminus of NHERF-2 showed very little interaction with β-catenin (lane 8), suggesting the affinity of the C terminus may not be strong enough to maintain interactions in vivo, or perhaps other binding partners for the C terminus of NHERF-2 prevent its association with β-catenin when the PDZ II domain is not present. Constructs of NHERF-2 containing PDZ II and at least part of the C terminus had a modest increase in affinity for β-catenin relative to PDZ II alone (Figure 3C, lanes 5, 6, and 7), suggesting that the C terminus of NHERF-2 may strengthen the interaction between β-catenin and the PDZ II domain of NHERF-2.
NHERF-2 Is Associated with Membrane-bound β-Catenin
β-Catenin performs at least two functions in cells. It is a component of the Wnt signaling pathway and as such is a soluble cytosolic protein. In addition, β-catenin is a component of the adherens junction, and in this capacity is tightly associated with E-cadherin and α-catenin. Junctional β-catenin is not a soluble protein but rather is membrane associated, and its extraction is dependent on detergent. To determine whether NHERF-2 is associated with β-catenin that is present in the cadherin/catenin complex, detergent extracts of membrane fractions of A431 and HT1080 cells, each expressing full-length exogenous NHERF-2, were immunoprecipitated using an antibody against NHERF-2 (32B6). A431 cells express E-cadherin and HT1080 cells express N-cadherin. Figure 3D shows that NHERF-2 coimmunoprecipitated not only with membrane-associated β-catenin but also with the other adherens junction proteins, E-cadherin, N-cadherin, and α-catenin, suggesting that NHERF-2 is capable of interacting with the pool of β-catenin that is in the cadherin/catenin complex.
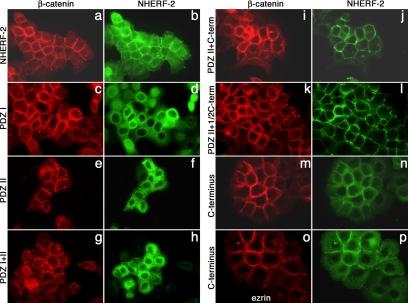
Immunofluorescence localization has been used to show that β-catenin is in cell–cell junctions in epithelial cells. To further substantiate that NHERF-2 is associated with β-catenin in junctions, we used immunofluorescence to colocalize the two proteins in A431 cells transfected with the various constructs of NHERF-2 (Figure 4). Figure 4, a and b, shows that transfected NHERF-2 localized at cell borders with β-catenin, which is indicative of adherens junction localization. There was additional staining in the cells suggesting that only a portion of NHERF-2 interacts with β-catenin. This is not surprising, considering the multiple functions ascribed to NHERF-2. Figure 4, c and d, shows that the construct containing only the first PDZ domain of NHERF-2 did not colocalize with β-catenin and was diffuse throughout the cytoplasm, which is in agreement with the binding assays presented above. As seen in Figure 4, e–l, all NHERF-2 constructs that contained the PDZ II domain colocalized with β-catenin at cell–cell borders. Figure 4, e–h, compares the staining pattern of β-catenin with that of NHERF-2 in cells expressing mutant proteins that lack the C terminus. These two mutant forms localized to cell–cell contacts, but they showed less cell border staining than full-length NHERF-2 (compare f and h to b).
Figure 4.
Colocalization of NHERF-2 and β-catenin. A431 cells expressing NHERF-2 and the constructs shown in Figure 2A were fixed on coverslips and stained for β-catenin (red), using rabbit anti-β-catenin and NHERF-2 (green) using 32B6 (against PDZ II) in b, f, h, j, and l, 9E10 in d and n, and rabbit anti-myc in p. A431 cells expressing the C terminus of NHERF-2 were stained for ezrin (red) in o.
As seen in Figure 4, i and j, NHERF-2 containing PDZ II + C terminus colocalized almost exclusively with β-catenin at cell borders. This is consistent with the protein-binding studies above showing that both PDZ II and the C terminus are required for highest affinity between the two molecules. Figure 4l shows the staining pattern of a NHERF-2 construct that contains the second PDZ domain and the C-terminal tail up through amino acid 280. The localization of this protein is similar to that of the PDZ II + C-terminus construct (j), suggesting that PDZ II and a region of the C terminus near PDZ II are sufficient for robust association with β-catenin. The very C terminus of NHERF-2 contains an ezrin-binding domain, and others have suggested that NHERF localizes to membranes via its association with ezrin (Yun et al., 1998). Figure 4, n and p, compares the staining pattern of a construct made up of only the C-terminal 90 amino acids of NHERF-2 to that of β-catenin (m) and ezrin (o). This construct lacks PDZ II but contains the ezrin-binding domain, confirming that binding to ezrin is sufficient to localize NHERF-2 to membranes. This construct is both membrane associated and diffuse, and, in addition to its interactions with membrane-associated ezrin, likely interacts with ezrin that is not at the membrane and/or with other binding partners. The PDZ II + 1/2 C-terminus construct, which does not include the ezrin-binding domain, is almost exclusively colocalized with β-catenin (l), indicating that NHERF-2 can be localized at the plasma membrane independently of its interaction with ezrin.
Interactions between β-Catenin and NHERF-2 Require a C-Terminal PDZ Binding Motif in β-Catenin
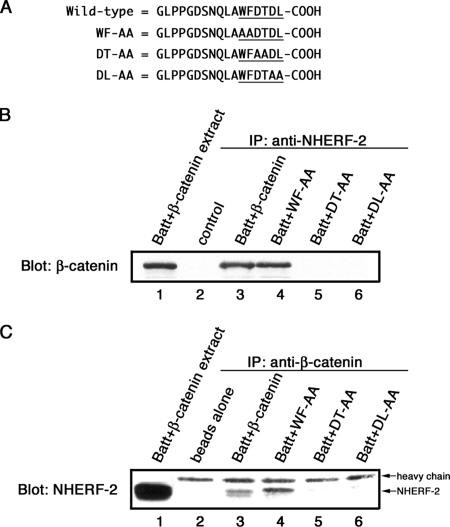
The very carboxy terminus of β-catenin contains a consensus PDZ-binding motif (DTDL). To examine a potential role for this sequence in binding to NHERF-2, a series of mutations were generated in the C terminus of β-catenin, and the protein was expressed in Batt cells. The Batt cell line is an ideal model for characterizing this interaction, because it expresses endogenous NHERF-2 but it does not express β-catenin. Successive dialanine mutations were made in the last six amino acids of β-catenin (WFDTDL) to generate three mutant forms: WF-AA, DT-AA, and DL-AA (Figure 5A). These mutant forms or wild-type β-catenin was expressed in Batt cells, extracts were made, and immunoprecipitations were done with antibodies against NHERF-2 (Figure 5B). Wild-type transfected β-catenin coimmunoprecipitated with endogenous NHERF-2 (lane 3). The WF-AA mutation did not prevent β-catenin from coimmunoprecipitating with NHERF-2 (lane 4), whereas replacing the DT or DL residues with two alanines, prevented NHERF-2 binding (lanes 5 and 6). The reciprocal immunoprecipitations were done with anti-β-catenin antibody conjugated to Sepharose (Figure 5C). It was necessary to conjugate the antibody to Sepharose to limit the amount of IgG heavy chain that would be present on the blot, because it nearly comigrates with NHERF-2 on SDS-PAGE. Endogenous NHERF-2 was associated with transfected, wild-type β-catenin (lane 3) as well as with the form containing the WF-AA mutation (lane 4). The β-catenin forms mutated at amino acids DT and DL were unable to coimmunoprecipitate NHERF-2 (lanes 5 and 6). The very C-terminal hydrophobic residue together with a serine/threonine at the −2 position is essential for class I PDZ interactions (Songyang et al., 1997). Thus, the data presented here suggest that β-catenin has a class I PDZ interaction domain that interacts with PDZ II of NHERF-2.
Figure 5.
β-catenin/NHERF-2 interaction requires the PDZ-binding motif of β-catenin. (A) Batt cells were transfected with wild-type β-catenin or β-catenin with the C-terminal mutations shown. (B) Extracts of transfected cells were immunoprecipitated with anti-NHERF-2 (32B6), resolved by SDS-PAGE, and immunoblotted with anti-β-catenin (15B8). (C) Transfected Batt cell extracts were immunoprecipitated with anti-β-catenin (15B8)-conjugated Sepharose and resolved by SDS-PAGE. 15B8-conjugated beads alone were loaded in lane 2 to indicate where 15B8 IgG heavy chain migrated in the gel. An immunoblot with anti-NHERF-2 (32B6) showed that endogenous NHERF-2 coimmunoprecipitated with transfected wild-type β-catenin (lane 3) and with β-catenin containing a WF-AA mutation (lane 4) but not the other two mutant forms of β-catenin (lanes 5 and 6).
NHERF-2 Colocalizes with Adherens Junction Proteins at Membrane Ruffles in HT1080 Cells
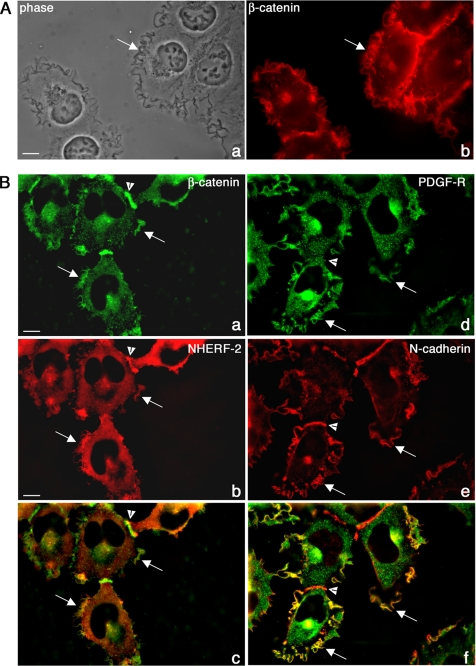
It is clear from the data presented above that NHERF-2 interacts with β-catenin in living cells; however, this does not tell us what function this complex plays. We know from previous reports that NHERFs interact with the PDGF receptor using their first PDZ domain. In addition, we know that activation of the PDGF receptor promotes cell motility in some cell types. HT1080 cells express low levels of the PDGF receptor and constitutively express PDGF (Gupta et al., 2001), which maintains the receptor in an active state. In addition, HT1080 cells express low levels of the NHERF-2 homologue NHERF-1, but they do not express NHERF-2. Finally, they express both N-cadherin and β-catenin. Thus, HT1080 cells provide an excellent model system to manipulate the expression of NHERF-2 and the level of PDGF-R to investigate their interactions with β-catenin and N-cadherin. In addition, HT1080 cells are highly motile with active membrane ruffles that are very obvious in phase microscopy, and surprisingly, they have N-cadherin and β-catenin prominently localized in these ruffles (Figure 6A, arrows). Thus HT1080 cells also serve as a model system for investigations into a possible role for a β-catenin/N-cadherin/NHERF/PDGF-R complex in cell motility.
Figure 6.
Characterization of membrane ruffles in HT1080 cells. (A) HT-1080 cells were grown on glass coverslips, processed for immunofluorescence, and stained for β-catenin. Phase (a) and fluorescence (b) micrographs are shown. (B) HT-1080 cells expressing NHERF-2 were processed for immunofluorescence and stained for β-catenin (rabbit β-catenin) and NHERF-2 (32B6). Images were deconvolved as described in Materials and Methods. β-Catenin (a) and NHERF-2 (b) were colocalized in junctions (arrowheads) and in membrane ruffles (arrows) in HT1080. A merged micrograph is shown in c. HT1080 cells expressing PDGF-Rβ were processed for immunofluorescence and costained for N-cadherin (e) and PDGF-Rβ (d), which colocalized in membrane ruffles (arrows). N-cadherin also localized to sites of cell–cell contact where PDGF-Rβ did not (arrowhead). A merged micrograph is shown in f.
Because it is not typical to see adherens junction molecules localized to free cell borders, we ruled out the possibility that this staining pattern was an “edge artifact” by staining HT-1080 cells for another junctional protein that we would not expect to find in membrane ruffles, α-v integrin. Supplemental Figure 1 shows that α-v integrin was not enriched in membrane ruffles and localized to focal contacts as expected, whereas β-catenin was enriched in the lamellipodia.
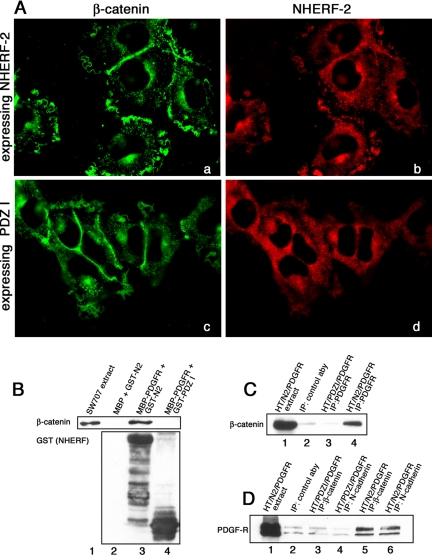
We expressed NHERF-2 and PDGF-Rβ in HT1080 cells using a retroviral expression system, and we showed that exogenously expressed NHERF-2 colocalized with β-catenin in junctions in HT1080 cells as we have shown above for A431 cells (Figure 6B, arrowheads). In addition, NHERF-2 colocalized with β-catenin in the lamellipodial structures (Figure 6B, arrows). Figure 6B, d, e, and f, shows that N-cadherin and PDGF-R are also in the lamellipodial structures, where they colocalize. Arrowheads point out cell–cell contacts, which are enriched in β-catenin, N-cadherin, and NHERF-2. Thus, N-cadherin, β-catenin, NHERF, and PDGF-R are all intensely localized in the prominent membrane ruffles found in HT1080 cells. If NHERF-2 were localized to membrane ruffles due to its interactions with β-catenin, we would predict that the NHERF-2 construct lacking the second PDZ domain would not be localized in ruffles. Figure 7A shows that full-length NHERF-2 colocalizes with β-catenin in membrane ruffles (a and b), whereas a construct of NHERF-2 truncated after the first PDZ domain is diffuse and does not colocalize with β-catenin in membrane ruffles (c and d).
Figure 7.
β-Catenin and N-cadherin form a complex with PDGF-Rβ and NHERF-2 in HT1080 cells. (A) HT1080 cells expressing NHERF-2 (a and b) or the first PDZ domain of NHERF-2 (c and d) were fixed on coverslips and costained for β-catenin (rabbit) and NHERF-2 using 33E2 against PDZ I. Deconvolved images are shown. (B) The C terminus of PDGF-Rβ fused to MBP or MBP alone were immobilized on amylose and incubated with SW707 extract in the presence of GST-wild type NHERF-2 (GST-N2) or GST-PDZ I domain of NHERF-2 (PDZ I). The reactions were resolved by SDS-PAGE and immunoblotted for β-catenin (top) or GST (bottom). (C) Extracts of HT1080 cells expressing PDGF-Rβ and full-length NHERF-2 (lanes 1, 2, and 4) or the first PDZ domain of NHERF-2 (lane 3) were immunoprecipitated with control antibody (lane 2) or anti-PDGF receptor (lanes 3 and 4), resolved by SDS-PAGE and immunoblotted for β-catenin. (D) Extracts of HT1080 cells expressing PDGF-Rβ and full-length NHERF-2 (lanes 1, 2, 5, and 6) or the first PDZ domain of NHERF-2 (lanes 3 and 4) were immunoprecipitated with control antibody (lane 2), antibodies against N-cadherin (lanes 4 and 6), or antibodies against β-catenin (lanes 3 and 5) and immunoblotted for PDGF-Rβ.
The interaction of NHERF-2 with β-catenin requires the second PDZ domain of NHERF-2, presumably leaving its first PDZ domain free to associate with other molecules. It has been shown that the first PDZ domain of NHERF-1 can associate with the C-terminal PDZ-binding motif of PDGF-Rβ (Maudsley et al., 2000). Thus, we proposed that NHERF-2 could form a bridging complex with PDGF-Rβ and β-catenin by using its two PDZ domains. To test this hypothesis, we prepared a construct similar to that used by Maudsley et al. (2000) encoding the C-terminal 53 amino acids of PDGF-Rβ, which comprises the PDZ interaction motif, and we tagged it at the N terminus with MBP. We immobilized the PDGF-Rβ fusion protein on amylose resin and incubated it with purified full-length GST-NHERF-2 in the presence of SW707 extract (containing β-catenin), washed the resin, resolved the bound proteins by SDS-PAGE, and immunoblotted for GST-NHERF and for β-catenin. Figure 7B, lane 3, shows that NHERF-2 bound to the PDGF-Rβ PDZ-binding motif and that β-catenin was bound to the full-length NHERF-2. When a purified construct of NHERF-2 that included the first PDZ I domain but not the second domain was used in the pull-down experiment in place of full-length NHERF-2, it was able to bind to the PDGF-Rβ on the resin, but it could not facilitate formation of a complex containing β-catenin (lane 4).
Next, we wanted to determine whether NHERF-2 could form a complex in vivo that included both β-catenin (via PDZ II) and PDGF-Rβ (via PDZ I). If NHERF-2 serves as a scaffold to link the N-cadherin/β-catenin complex to PDGF-Rβ, we would expect that expression of the PDZ I domain alone of NHERF-2, which can bind to PDGF-R but cannot bind to β-catenin, would act as a dominant negative and block PDGF-Rβ interactions with β-catenin. When we exogenously expressed NHERF-2 together with PDGF-Rβ in HT1080 cells and immunoprecipitated cell extracts with antibodies against PDGF-Rβ, we saw that β-catenin was in the complex (Figure 7C, lane 4). This association was lost in cells expressing the NHERF-2 mutant containing only the first PDZ domain (Figure 7C, lane 3). In addition, PDGF-Rβ coimmunoprecipitated with N-cadherin and with β-catenin in cells expressing PDGF-Rβ and full-length NHERF-2 (Figure 7D, lanes 5 and 6) but not in cells expressing the PDZ I domain (Figure 7D, compare lanes 3 and 4 with lane 2, which was immunoprecipitated with control antibody).
The N-Cadherin/PDGF Receptor Complex Contributes to HT1080 Cell Migration
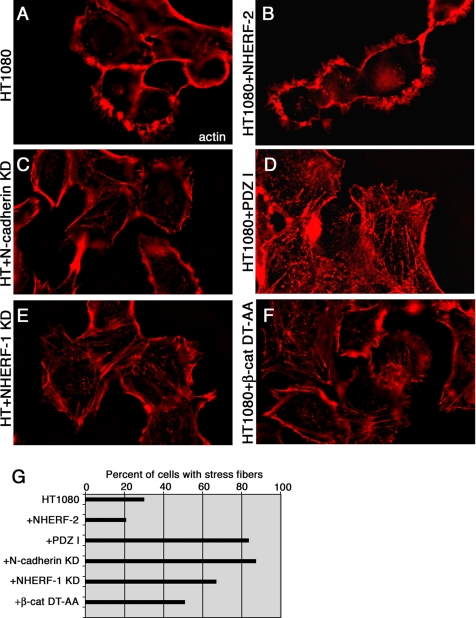
To determine whether the NHERF-2/N-cadherin/β-catenin/ PDGF receptor complex plays a role in cell motility, we examined actin cytoskeleton organization and cell motility in cells overexpressing an NHERF-2 mutant and in cells knocked down for N-cadherin or endogenous NHERF-1. Using immunofluorescence, we found that HT1080 cells expressing shRNA against endogenous NHERF-1 (Figure 8E) or shRNA against N-cadherin (Figure 8C) had more stress fibers and less actin localized in lamellipodia than did parental HT1080 cells (Figure 8A) or cells expressing full-length NHERF-2 (Figure 8B), suggesting they may be less motile. In addition, cells expressing a β-catenin mutant unable to bind to NHERF (Figure 8F), and cells expressing the PDZ I domain of NHERF-2, which cannot bind to β-catenin (Figure 8D) had increased stress fibers, compared with parental HT1080 cells. Supplemental Figure 2A (lanes 1 and 2) shows that HT1080 cells expressing shRNA against N-cadherin had significant knockdown of N-cadherin expression, and Supplemental Figure 2B (lanes 1 and 2) shows that HT1080 cells expressing shRNA against NHERF-1 had significant knockdown of NHERF-1 expression. Supplemental Figure 2C shows that the expression level of the mutant β-catenin was approximately twice that of endogenous β-catenin; thus, it was likely sufficient to interfere with formation of a complex among β-catenin, NHERF, and PDGF-R.
Figure 8.
Disruption of the scaffolding activity of NHERF induces formation of actin stress fibers. Parental HT1080 cells (A), HT1080 cells expressing exogenous NHERF-2 (B), HT1080 cells expressing shRNA against N-cadherin (HT + N-cadherin KD; C), HT1080 cells expressing the PDZ I domain of NHERF-2 that cannot bind to β-catenin (D), HT1080 cells expressing shRNA against endogenous NHERF1 (HT + NHERF-1 KD; E), and HT1080 cells expressing the β-catenin mutant unable to bind to NHERF (β-cat DT-AA; F) were fixed on coverslips and stained with rhodamine-phalloidin to visualize filamentous actin. (G) Three hundred cells were evaluated, and the percentage of cells with stress fibers was graphed.
We examined 300 cells each of the parental HT1080 cells (Figure 8A), cells expressing shRNA against N-cadherin (Figure 8C), cells expressing shRNA against endogenous NHERF1 (Figure 8E), cells expressing exogenous NHERF-2 (Figure 8B), cells expressing the β-catenin mutant unable to bind to NHERF (β-cat DT-AA; Figure 8F), and cells expressing the PDZ I domain of NHERF-2, which cannot bind to β-catenin (Figure 8D), and we evaluated them microscopically for the presence of actin in stress fibers. Because lamellipodial actin and stress fiber actin are not in the same plane of focus in the microscope, we collected Z stacks of each cell population and created a three-dimensional stack so the reader can view the entire cell, just as we saw it in the microscope. Supplemental Movie 1 shows the three-dimensional stack of HT1080 cells expressing NHERF-2 as an example of cells with few stress fibers, and Supplemental Movie 2 shows the three-dimensional stack of HT1080 cells expressing shRNA against NHERF-1 as an example of cells with numerous stress fibers. The percentage of cells with stress fibers was calculated and graphed (Figure 8G). There was an increase in the percentage of cells with stress fibers in all cases where the β-catenin/N-cadherin/NHERF complex was disrupted. Approximately 20–30% of parental HT1080 cells or NHERF-2–expressing HT1080 cells had stress fibers, whereas >80% of cells expressing the PDZ I domain of NHERF-2 had stress fibers; 70–90% of cells knocked down for NHERF I or knocked down for N-cadherin, respectively, had stress fibers; and ∼50% of cells expressing β-catenin that could not bind to NHERF had stress fibers.
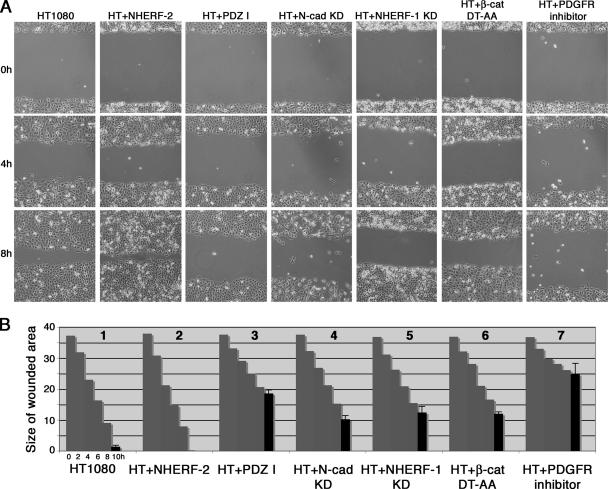
To determine whether the observed changes in the actin cytoskeleton translated to changes in cell motility, we performed wound-healing assays (Figure 9). Confluent monolayers of parental HT1080 cells, HT1080 cells expressing NHERF-2, HT1080 cells expressing the PDZ I domain of NHERF-2, HT1080 cells knocked down for N-cadherin, HT1080 cells knocked down for NHERF-1, HT1080 cells expressing β-catenin that cannot bind to NHERF (β-cat DT-AA), and parental HT1080 cells treated with a PDGF-R inhibitor were wounded and photographed immediately after producing the wound (0 h) and at 2, 4, 6, 8, and 10 h postwounding. Figure 9A shows phase micrographs of the 0-, 4-, and 8-h time points. Motility was quantified by measuring the remaining cleared area and the data are represented as a graph (Figure 9B). HT1080 cells expressing shRNA against N-cadherin or shRNA against NHERF-1 were not able to fill the wounded area as quickly as parental HT1080 cells (Figure 9B, compare graphs 4 and 5, respectively, with graph 1). Activation of PDGF-Rβ in HT1080 contributes to cell motility (Gupta et al., 2001), and, as expected, treatment with a PDGF-R kinase inhibitor greatly reduced motility in this assay (compare graph 7 with graph 1). Cells expressing the NHERF-2 mutant containing only the first PDZ domain that cannot bind to β-catenin, or the β-catenin mutant that cannot bind to NHERF showed decreased ability to migrate into the wounded area (compare graphs 3 and 6, respectively, with graph 1). Together, the data presented in Figures 8 and 9 show that perturbing the formation of an N-cadherin/β-catenin/NHERF/PDGF-R complex perturbs the actin cytoskeleton and reduces cell motility.
Figure 9.
The N-cadherin/β-catenin/NHERF/PDGF-R complex contributes to HT1080 cell migration. (A) Confluent monolayers of parental HT1080 cells, HT1080 cells expressing exogenous NHERF-2, (HT + NHERF-2), HT1080 cells expressing the PDZ I domain of NHERF-2 that cannot bind to β-catenin (HT + PDZ I), HT1080 cells expressing shRNA against N-cadherin (HT + N-cad KD), HT1080 cells expressing shRNA against endogenous NHERF1 (HT + NHERF-1 KD), HT1080 cells expressing the β-catenin mutant unable to bind to NHERF (HT + β-cat DT-AA) or parental HT1080 cells treated with PDGF-R inhibitor (HT + PDGF-R inhibitor) were scratched with a sterile pipette tip and allowed to migrate into the wound for 10 h. (B) The above-mentioned assays were quantified by measuring the amount of denuded dish remaining at the specified times. Three independent assays were done and the SD calculated for the final time point (10 h).
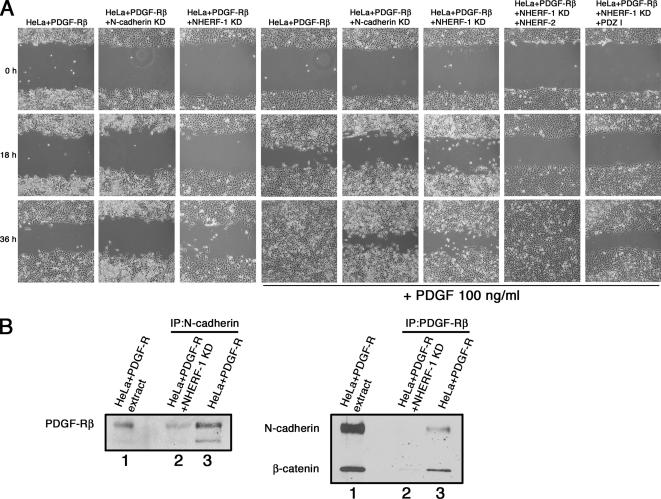
To ensure we were not looking at a phenomenon specific to HT1080 cells, we sought to determine whether a complex between N-cadherin/β-catenin/NHERF/PDGF-R influences cell motility in other cells. We used HeLa cells that express N-cadherin, β-catenin, and NHERF-1, but, unlike HT0180 cells, these cells do not have constitutively activated PDGF-R. When HeLa cells were infected with a retrovirus encoding PDGF-Rβ, they displayed PDGR-dependent motility that was diminished upon expression of shRNA against N-cadherin or NHERF-1 (Figure 10A). PDGF-dependent motility was rescued in NHERF-1 knock-down cells by expressing full-length NHERF-2 but not by expressing the mutant containing only the first PDZ domain. This indicates that NHERFs are functionally redundant in PDGF signaling. PDGF-R coimmunoprecipitated with N-cadherin from cells expressing NHERF-1 but not from cells knocked down for NHERF-1. When the reciprocal immunoprecipitations were done with antibodies against PDGF-R, both N-cadherin and β-catenin coimmunoprecipitated with PDGF-R from cells expressing NHERF-1 but not from cells knocked down for NHERF-1 (Figure 10B). Supplemental Figure 2A, lanes 3 and 4, shows that HeLa cells expressing shRNA against N-cadherin had significant knockdown of N-cadherin expression, and Supplemental Figure 2B, lanes 3 and 4, shows that HeLa cells expressing shRNA against NHERF-1 had significant knockdown of NHERF-1 expression. Together, these results indicate that NHERF forms a bridge between the N-cadherin/β-catenin complex and PDGF-R to regulate cell motility in HeLa cells, much like it does in HT1080 cells.
Figure 10.
The N-cadherin/β-catenin/NHERF/PDGF-R complex contributes to HeLa cell migration. (A) HeLa cells expressing PDGF-Rβ, HeLa cells expressing PDGF-Rβ together with shRNA against N-cadherin (N-cadherin KD), or HeLa cells expressing PDGF-Rβ together with shRNA against NHERF-1 (NHERF-1 KD) were serum starved for 24 h, scratched with a sterile pipette tip, and treated with (second 3 panels) or without (first 3 panels) PDGF-BB ligand. The right two panels show HeLa cells expressing PDGF-Rβ together with shRNA against NHERF-1 and full-length NHERF-2, or PDGF-Rβ together with shRNA against NHERF-1 and the NHERF-2 PDZ I domain alone. Cells were allowed to migrate into the denuded area, and phase micrographs taken at 0, 18, and 36 h. (B) HeLa cells expressing PDGF-Rβ were extracted, immunoprecipitated with antibodies against N-cadherin, and immunoblotted for PDGF-Rβ (left) or immunoprecipitated with antibodies against PDGF-Rβ and immunoblotted for N-cadherin and β-catenin (right). On both gels, lane 1 is cell extract and lanes 2 and 3 are the immunoprecipitation reactions.
DISCUSSION
PDZ domains typically interact with specific C-terminal sequences on their partner proteins. Here, we show that the second PDZ domain of NHERF-2 binds to the class I consensus motif found at the C terminus of β-catenin. We also show that β-catenin weakly interacts with the C terminus of NHERF-2 in vitro and that the binding affinity of β-catenin with NHERF-2 is mediated by PDZ II and a portion of the C terminus. Our results are similar to another study, which concluded that NHERF-2 association with CFTR requires both the PDZ II domain and the C terminus (Yun et al., 1998). This study showed that the second PDZ domain of NHERF-2 alone was not sufficient to bind its partner CFTR. In contrast, we show that the NHERF-2 PDZ II domain is able to associate with β-catenin and that addition of at least a portion of the C-terminal domain enhances biochemical interaction and colocalization in A431 cells. This region of interaction is consistent with our original phage display results, in which the isolated NHERF-2 clone encoded a portion of the protein including the second PDZ domain and part of the C-terminal region.
NHERF-2 has been called by other names in the literature, including E3KARP, SIP-1, and TKA-1. Na+/H+ exchanger type 3 kinase A regulatory protein (E3KARP) is another name for authentic NHERF-2 (Sun et al., 2000). SRY interacting protein (SIP-1; ACC U82108) is an alternatively spliced form of NHERF-2, missing 11 amino acids (286–296) near the C terminus, but N-terminal to the ERM-binding site. SIP-1was shown to associate with the product of SRY during testis development in human embryogenesis, and it was identified as a SRY binding partner in a yeast-two hybrid screen against a human fetal cDNA library. SIP-1 was subsequently shown to localize in the nucleus with SRY in a human embryonic cell line and in gonadal tissue from a 7-wk-old male fetus. It is unclear whether this alternatively spliced form of NHERF-2 is expressed in other cell types (Poulat et al., 1997), but several expressed sequence tags containing this 11-amino acid deletion are in the database. TKA-1 (ACC Z50150) was described as a tyrosine kinase-activating protein of 450 amino acids that activates PDGF signaling. The first 307 amino acids of TKA-1 are identical to NHERF-2, except that it is missing amino acid 51 of NHERF-2. However, the remaining amino acids are completely different. The TKA-1 nucleotide sequence has a single nucleotide deletion relative to both NHERF-2 (ACC AF035771) and the human genome sequence. The latter comparison shows the deletion is not at an exon boundary. These data raise the possibility that the additional C-terminal sequence in TKA-1 may not represent an authentic isoform of NHERF-2.
Others have shown that β-catenin associates with PDZ domain-containing proteins, including the NHERF-2 homologue NHERF-1 (Shibata et al., 2003). This study indicated that NHERF-1 functions as a partner with β-catenin in the nucleus to enhance β-catenin transcriptional activity. We did not expect NHERF-2 to play a role in β-catenin–mediated transcription, because we saw colocalization of NHERF-2 with β-catenin at the membrane but not in the nucleus. Also, it has been shown that cells expressing a β-catenin mutant lacking the C-terminal PDZ-binding motif have no change in β-catenin transcriptional activity (Kanamori et al., 2003). The PDZ containing proteins MAGI-1 and LIN-7 have been shown to interact with the C terminus of β-catenin (Dobrosotskaya and James, 2000; Perego et al., 2000). Both of these proteins are thought to function as scaffolds and to play a role in cell polarity. NHERF proteins also function as scaffolds between membrane and cytoplasmic proteins. Yun et al. (1998) and Weinman et al. (2006) showed distinct membrane localization of NHERF-2 in the Chinese hamster fibroblast cell line PS120, and they suggested that NHERF-2 functions as a scaffold between NHE3 and ezrin. They showed colocalization of NHERF-2 with ezrin at the plasma membrane and mapped the binding domain to the very C terminus of NHERF-2. Here, we show that NHERF-2 localizes to the membrane in A431 cells where it colocalizes with β-catenin in the adherens junction and that this localization does not require the ezrin-binding domain.
Because full-length NHERF-2 contains two PDZ domains, it could associate with two different molecules. Interestingly, immunofluorescence shows that NHERF-2 constructs containing the second PDZ domain and at least a portion of the C terminus localize distinctly with β-catenin at the plasma membrane. NHERF-2 constructs containing the first PDZ domain, i.e., full-length NHERF-2 and the construct containing both PDZ domains without the C terminus show some diffuse staining in areas other than cell borders. It is likely that the first PDZ domain of NHERF-2 has cellular binding partners other than β-catenin, and some of the exogenously expressed protein associates with these other partners. Without competition, the second PDZ domain and the C terminus bind strongly to β-catenin in binding assays and localize distinctly at the membrane.
In HT1080 cells, which express PDGF receptors, it is likely that NHERF-2 associates with the receptor at the first PDZ domain, leaving the second domain free to bind other partners. To date, no complex of proteins has been identified that includes PDGF-R bound to the NHERF PDZ I domain, and at the same time, a second binding partner bound to the NHERF PDZ II domain. Here, we show that β-catenin binds to the second PDZ domain of NHERF-2 both in vitro and in cells and that N-cadherin and β-catenin are present in an immune complex with PDGF-Rβ. A construct of NHERF-2 that is unable to bind β-catenin prevents such coimmunoprecipitations, indicating for the first time a role for NHERFs in scaffolding PDGF-R to other membrane-associated complexes.
It has been shown by others that NHERF-1 associates with PDGF-R to regulate its function. Mouse embryo fibroblasts (MEFs) expressing PDGF-Rβ mutated in the PDZ binding domain, which is unable to associate with NHERF, showed reduced cell spreading and motility compared with cells expressing wild-type PDGF-Rβ (James et al., 2004). Another study showed that NHERF interaction with the PDGF-R is ligand dependent and perturbs PDGF-R–mediated actin cytoskeletal reorganization in porcine aortic endothelial cells, presumably by inhibiting Rho GTPases such as Rac and Cdc42 (Demoulin et al., 2003). Notably, the latter study showed no change in chemotaxis when cells expressing a PDGF-Rβ mutant that cannot bind NHERF were plated on collagen or fibronectin. We show here that disruption of the cadherin/β-catenin/NHERF complex by down regulating N-cadherin or NHERF-1, or expressing forms of NHERF-2 or β-catenin that are uncoupled from one another, promotes stress fiber formation and reduces cell motility, suggesting that this complex plays a role in actin reorganization and cell motility. Our model agrees with the studies done in MEFs showing decreased motility in cells expressing a NHERF uncoupled PDGF-R mutant and suggests that NHERF-2, PDGF-Rβ, N-cadherin, and β-catenin form a macromolecular complex in membrane ruffles that contributes to cell motility. Thus, one-way N-cadherin may promote cell motility is by modulating PDGF-R signaling. Recently, the Reynolds laboratory showed a connection between adherens junction molecules and PDGF-mediated actin reorganization (Wildenberg et al., 2006). In their study, p120 catenin and N-cadherin are required for dorsal circular ruffle formation in NIH3T3 cells treated with PDGF. This group showed that p120 is required for recruitment of p190RhoGAP and inhibition of RhoA signaling. It will be interesting to determine what roles p120 catenin and p190RhoGAP play in the NHERF-dependent processes described in the current study.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jill Nieset and Jennifer Oiler for excellent technical assistance. This work was supported by grants DE-12308 and GM-51188 from the National Institutes of Health (to K.R.J. and M.J.W., respectively). C.T. was supported by the Ruth C. Kirschstein National Research Service Award T32CA09476-14.
Abbreviations used:
- NHERF
Na+/H+ exchanger regulatory factor
- PDZ
PSD-95/Dlg/ZO-1
- PDGF-R
platelet-derived growth factor receptor.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-10-0960) on January 17, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Ades E. W., Candal F. J., Swerlick R. A., George V. G., Summers S., Bosse D. C., Lawley T. J. HMEC-1, establishment of an immortalized human microvascular endothelial cell line. J. Invest. Dermatol. 1992;99:683–690. doi: 10.1111/1523-1747.ep12613748. [DOI] [PubMed] [Google Scholar]
- Bienz M. beta-Catenin: a pivot between cell adhesion and Wnt signalling. Curr. Biol. 2005;15:R64–R67. doi: 10.1016/j.cub.2004.12.058. [DOI] [PubMed] [Google Scholar]
- Brembeck F. H., Rosario M., Birchmeier W. Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr. Opin. Genet. Dev. 2006;16:51–59. doi: 10.1016/j.gde.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Demoulin J. B., Seo J. K., Ekman S., Grapengiesser E., Hellman U., Ronnstrand L., Heldin C. H. Ligand-induced recruitment of Na+/H+-exchanger regulatory factor to the PDGF (platelet-derived growth factor) receptor regulates actin cytoskeleton reorganization by PDGF. Biochem. J. 2003;376:505–510. doi: 10.1042/BJ20030385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrosotskaya I. Y., James G. L. MAGI-1 interacts with beta-catenin and is associated with cell-cell adhesion structures. Biochem. Biophys. Res. Commun. 2000;270:903–909. doi: 10.1006/bbrc.2000.2471. [DOI] [PubMed] [Google Scholar]
- Falcone D., Andrews D. W. Both the 5′ untranslated region and the sequences surrounding the start site contribute to efficient initiation of translation in vitro. Mol. Cell Biol. 1991;11:2656–2664. doi: 10.1128/mcb.11.5.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury-Feith J., Kheuang L., Zeng L., Bignon J., Boutin C., Monnet I., Jaurand M. C. Human malignant mesothelial cells: variability of ultrastructural features in established and nude mice transplanted cell lines. J. Pathol. 1995;177:209–215. doi: 10.1002/path.1711770215. [DOI] [PubMed] [Google Scholar]
- Gottardi C. J., Gumbiner B. M. Adhesion signaling: how beta-catenin interacts with its partners. Curr. Biol. 2001;11:R792–R794. doi: 10.1016/s0960-9822(01)00473-0. [DOI] [PubMed] [Google Scholar]
- Grignani F., Kinsella T., Mencarelli A., Valtieri M., Riganelli D., Lanfrancone L., Peschle C., Nolan G. P., Pelicci P. G. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 1998;58:14–19. [PubMed] [Google Scholar]
- Gupta S., Stuffrein S., Plattner R., Tencati M., Gray C., Whang Y. E., Stanbridge E. J. Role of phosphoinositide 3-kinase in the aggressive tumor growth of HT1080 human fibrosarcoma cells. Mol. Cell Biol. 2001;21:5846–5856. doi: 10.1128/MCB.21.17.5846-5856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan R. B., Phillips G. R., Qiao R. F., Norton L., Aaronson S. A. Exogenous expression of N-cadherin in breast cancer cells induces cell migration, invasion, and metastasis. J. Cell Biol. 2000;148:779–790. doi: 10.1083/jcb.148.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin C. H., Ostman A., Ronnstrand L. Signal transduction via platelet-derived growth factor receptors. Biochim. Biophys. Acta. 1998;1378:F79–F113. doi: 10.1016/s0304-419x(98)00015-8. [DOI] [PubMed] [Google Scholar]
- Hirt R. P., Poulain-Godefroy O., Billotte J., Kraehenbuhl J. P., Fasel N. Highly inducible synthesis of heterologous proteins in epithelial cells carrying a glucocorticoid-responsive vector. Gene. 1992;111:199–206. doi: 10.1016/0378-1119(92)90687-k. [DOI] [PubMed] [Google Scholar]
- Ireton R. C., et al. A novel role for p120 catenin in E-cadherin function. J. Cell Biol. 2002;159:465–476. doi: 10.1083/jcb.200205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S., Carey T. E., Wolf G. T., Wheelock M. J., Johnson K. R. Expression of N-cadherin by human squamous carcinoma cells induces a scattered fibroblastic phenotype with disrupted cell-cell adhesion. J. Cell Biol. 1996;135:1643–1654. doi: 10.1083/jcb.135.6.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James M. F., Beauchamp R. L., Manchanda N., Kazlauskas A., Ramesh V. A NHERF binding site links the betaPDGFR to the cytoskeleton and regulates cell spreading and migration. J. Cell Sci. 2004;117:2951–2961. doi: 10.1242/jcs.01156. [DOI] [PubMed] [Google Scholar]
- Johnson K. R., Lewis J. E., Li D., Wahl J., Soler A. P., Knudsen K. A., Wheelock M. J. P- and E-cadherin are in separate complexes in cells expressing both cadherins. Exp. Cell Res. 1993;207:252–260. doi: 10.1006/excr.1993.1191. [DOI] [PubMed] [Google Scholar]
- Kanamori M., Sandy P., Marzinotto S., Benetti R., Kai C., Hayashizaki Y., Schneider C., Suzuki H. The PDZ protein tax-interacting protein-1 inhibits beta-catenin transcriptional activity and growth of colorectal cancer cells. J. Biol. Chem. 2003;278:38758–38764. doi: 10.1074/jbc.M306324200. [DOI] [PubMed] [Google Scholar]
- Karthikeyan S., Leung T., Ladias J. A. Structural basis of the Na+/H+ exchanger regulatory factor PDZ1 interaction with the carboxyl-terminal region of the cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 2001;276:19683–19686. doi: 10.1074/jbc.C100154200. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamprecht G., Weinman E. J., Yun C. H. The role of NHERF and E3KARP in the cAMP-mediated inhibition of NHE3. J. Biol. Chem. 1998;273:29972–29978. doi: 10.1074/jbc.273.45.29972. [DOI] [PubMed] [Google Scholar]
- Leibovitz A., Stinson J. C., McCombs W. B., 3rd, McCoy C. E., Mazur K. C., Mabry N. D. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36:4562–4569. [PubMed] [Google Scholar]
- Maeda M., Johnson K. R., Wheelock M. J. Cadherin switching: essential for behavioral but not morphological changes during an epithelium-to-mesenchyme transition. J. Cell Sci. 2005;118:873–887. doi: 10.1242/jcs.01634. [DOI] [PubMed] [Google Scholar]
- Maudsley S., Zamah A. M., Rahman N., Blitzer J. T., Luttrell L. M., Lefkowitz R. J., Hall R. A. Platelet-derived growth factor receptor association with Na(+)/H(+) exchanger regulatory factor potentiates receptor activity. Mol. Cell Biol. 2000;20:8352–8363. doi: 10.1128/mcb.20.22.8352-8363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman M. T., Prudoff R. S., Johnson K. R., Wheelock M. J. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J. Cell Biol. 1999;147:631–644. doi: 10.1083/jcb.147.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego C., Vanoni C., Massari S., Longhi R., Pietrini G. Mammalian LIN-7 PDZ proteins associate with beta-catenin at the cell-cell junctions of epithelia and neurons. EMBO J. 2000;19:3978–3989. doi: 10.1093/emboj/19.15.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulat F., Barbara P. S., Desclozeaux M., Soullier S., Moniot B., Bonneaud N., Boizet B., Berta P. The human testis determining factor SRY binds a nuclear factor containing PDZ protein interaction domains. J. Biol. Chem. 1997;272:7167–7172. doi: 10.1074/jbc.272.11.7167. [DOI] [PubMed] [Google Scholar]
- Reczek D., Bretscher A. The carboxyl-terminal region of EBP50 binds to a site in the amino-terminal domain of ezrin that is masked in the dormant molecule. J. Biol. Chem. 1998;273:18452–18458. doi: 10.1074/jbc.273.29.18452. [DOI] [PubMed] [Google Scholar]
- Rudiger M., Jockusch B. M., Rothkegel M. Epitope tag-antibody combination useful for the detection of protein expression in prokaryotic and eukaryotic cells. Biotechniques. 1997;23:96–97. doi: 10.2144/97231bm20. [DOI] [PubMed] [Google Scholar]
- Sacco P. A., McGranahan T. M., Wheelock M. J., Johnson K. R. Identification of plakoglobin domains required for association with N-cadherin and alpha-catenin. J. Biol. Chem. 1995;270:20201–20206. doi: 10.1074/jbc.270.34.20201. [DOI] [PubMed] [Google Scholar]
- Sheffield P., Garrard S., Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- Shenolikar S., Weinman E. J. NHERF: targeting and trafficking membrane proteins. Am. J. Physiol. 2001;280:F389–F395. doi: 10.1152/ajprenal.2001.280.3.F389. [DOI] [PubMed] [Google Scholar]
- Shibata T., Chuma M., Kokubu A., Sakamoto M., Hirohashi S. EBP50, a beta-catenin-associating protein, enhances Wnt signaling and is over-expressed in hepatocellular carcinoma. Hepatology. 2003;38:178–186. doi: 10.1053/jhep.2003.50270. [DOI] [PubMed] [Google Scholar]
- Songyang Z., Fanning A. S., Fu C., Xu J., Marfatia S. M., Chishti A. H., Crompton A., Chan A. C., Anderson J. M., Cantley L. C. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- Sun F., Hug M. J., Lewarchik C. M., Yun C. H., Bradbury N. A., Frizzell R. A. E3KARP mediates the association of ezrin and protein kinase A with the cystic fibrosis transmembrane conductance regulator in airway cells. J. Biol. Chem. 2000;275:29539–29546. doi: 10.1074/jbc.M004961200. [DOI] [PubMed] [Google Scholar]
- Suyama K., Shapiro I., Guttman M., Hazan R. B. A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor. Cancer Cell. 2002;2:301–314. doi: 10.1016/s1535-6108(02)00150-2. [DOI] [PubMed] [Google Scholar]
- Voltz J. W., Weinman E. J., Shenolikar S. Expanding the role of NHERF, a PDZ-domain containing protein adapter, to growth regulation. Oncogene. 2001;20:6309–6314. doi: 10.1038/sj.onc.1204774. [DOI] [PubMed] [Google Scholar]
- Wahl J. K., 3rd. Generation of monoclonal antibodies specific for desmoglein family members. Hybrid Hybridomics. 2002;21:37–44. doi: 10.1089/15368590252917629. [DOI] [PubMed] [Google Scholar]
- Wahl J. K., 3rd, Kim Y. J., Cullen J. M., Johnson K. R., Wheelock M. J. N-cadherin-catenin complexes form prior to cleavage of the proregion and transport to the plasma membrane. J. Biol. Chem. 2003;278:17269–17276. doi: 10.1074/jbc.M211452200. [DOI] [PubMed] [Google Scholar]
- Wang S., Raab R. W., Schatz P. J., Guggino W. B., Li M. Peptide binding consensus of the NHE-RF-PDZ1 domain matches the C-terminal sequence of cystic fibrosis transmembrane conductance regulator (CFTR) FEBS Lett. 1998;427:103–108. doi: 10.1016/s0014-5793(98)00402-5. [DOI] [PubMed] [Google Scholar]
- Weinman E. J., Hall R. A., Friedman P. A., Liu-Chen L. Y., Shenolikar S. The association of NHERF adaptor proteins with g protein-coupled receptors and receptor tyrosine kinases. Annu. Rev. Physiol. 2006;68:491–505. doi: 10.1146/annurev.physiol.68.040104.131050. [DOI] [PubMed] [Google Scholar]
- Wildenberg G. A., Dohn M. R., Carnahan R. H., Davis M. A., Lobdell N. A., Settleman J., Reynolds A. B. p120-catenin and p190RhoGAP regulate cell-cell adhesion by coordinating antagonism between Rac and Rho. Cell. 2006;127:1027–1039. doi: 10.1016/j.cell.2006.09.046. [DOI] [PubMed] [Google Scholar]
- Yun C. H., Lamprecht G., Forster D. V., Sidor A. NHE3 kinase A regulatory protein E3KARP binds the epithelial brush border Na+/H+ exchanger NHE3 and the cytoskeletal protein ezrin. J. Biol. Chem. 1998;273:25856–25863. doi: 10.1074/jbc.273.40.25856. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.