Abstract
Similar to their human counterparts, the Drosophila Rbf1 and Rbf2 Retinoblastoma family members control cell cycle and developmentally regulated gene expression. Increasing evidence suggests that Rbf proteins rely on multiprotein complexes to control target gene transcription. We show here that the developmentally regulated COP9 signalosome (CSN) physically interacts with Rbf2 during embryogenesis. Furthermore, the CSN4 subunit of the COP9 signalosome co-occupies Rbf target gene promoters with Rbf1 and Rbf2, suggesting an active role for the COP9 signalosome in transcriptional regulation. The targeted knockdown of individual CSN subunits leads to diminished Rbf1 and Rbf2 levels and to altered cell cycle progression. The proteasome-mediated destruction of Rbf1 and Rbf2 is increased in cells and embryos with diminished COP9 activity, suggesting that the COP9 signalosome protects Rbf proteins during embryogenesis. Previous evidence has linked gene activation to protein turnover via the promoter-associated proteasome. Our findings suggest that Rbf repression may similarly involve the proteasome and the promoter-associated COP9 signalosome, serving to extend Rbf protein lifespan and enable appropriate programs of retinoblastoma gene control during development.
INTRODUCTION
In humans, the Retinoblastoma tumor suppressor protein (RB) and its related family members, p107 and p130, play important roles in coordinating cell cycle progression by controlling patterns of gene expression during proliferation (reviewed in Mulligan and Jacks, 1998; Classon and Dyson, 2001). Much interest has focused on the function of RB family members because the gene encoding RB is mutated in a wide variety of human tumors (Sellers and Kaelin, 1997; Nevins, 2001; Classon and Harlow, 2002). Although p107 and p130 share extensive similarities with RB, the p107 and p130 loci are infrequently mutated during tumorigenesis (Paggi et al., 1996), indicating that RB family members can perform specialized functions during normal cellular growth, division, and differentiation as well as during deregulated growth during cancer progression. Drosophila melanogaster has two retinoblastoma homologues, Rbf1 and Rbf2, which regulate cell cycle–specific and developmental genes (Dimova et al., 2003). Rbf1 appears to play a dominant role in this process; however, Rbf2 is found at many of the same promoters and may play a synergistic role in governing appropriate developmental patterns (Stevaux et al., 2005).
The function of RB family members in repression is best understood through their involvement with the E2F transcriptional activator family of proteins (Frolov and Dyson, 2004), although RB also plays important roles in growth control through E2F-independent pathways (Hirsch et al., 2000, 2004). RB may repress transcription by direct occlusion of the E2F activation domain or by recruitment of transcriptional corepressor proteins that directly repress transcription and/or modify chromatin structure at the promoter (Harbour and Dean, 2000). RB cofactors implicated in these processes include histone deactylases and methyltransferases, SWI/SNF chromatin remodeling machinery, Polycomb complexes, and DNA methyltransferases (Morris and Dyson, 2001). In Drosophila, both Rbf1 and Rbf2 can associate with a Myb-containing complex called dREAM (Korenjak et al., 2004; Lewis et al., 2004), which was previously demonstrated to play roles in both activation and repression of developmentally regulated DNA replication (Beall et al., 2002). Components of the dREAM complex are also involved in vulval cell fate in Caenorhabditis elegans (Korenjak et al., 2004), consistent with a role for RB in development. The plethora and diversity of factors demonstrated to associate with RB family members indicate that distinct mechanisms of repression may be invoked during different stages of the cell cycle and in gene-specific and developmental stage–specific patterns.
To understand the function of RB proteins during development, endogenous Rbf2 was purified from Drosophila embryos to identify associated proteins. This analysis revealed a previously uncharacterized association between Rbf2 and the developmentally regulated COP9 signalosome. The COP9 signalosome was first identified in Arabidopsis as a repressor of light-induced development and is composed of eight subunits (CSN1-8) that are highly conserved across plant and animal kingdoms (Wei and Deng, 1992, 2003). The COP9 signalosome was previously linked to the Rbf pathway through its regulation of cyclin E levels (Doronkin et al., 2003), and targeted reduction of Drosophila COP9 signalosome subunits by RNA interference (RNAi) results in defects in G1 progression, indicating a major role for this complex in governing cell cycle progression (Bjorklund et al., 2006). Our data indicate that the COP9 signalosome is involved at multiple steps in cell cycle control with a direct role in governing Rbf1 and Rbf2 stability during development.
MATERIALS AND METHODS
Purification and Identification of Rbf2-associated Proteins
Rbf2 was immunoprecipitated from nuclear extracts prepared from 0- to 12-h-old embryos. Five milliliters of rabbit α-Rbf2 or preimmune serum was covalently cross-linked to 5 ml of protein G agarose beads and incubated with 5 ml of nuclear extracts for 2 h at room temperature. The beads were washed six times in HEMGT-150 (25 mM HEPES, pH 7.9, 0.1 mM EDTA, 12.5 mM MgCl2, 5% glycerol, 0.1% Tween-20, 150 mM NaCl) containing protease inhibitors and dithiothreitol (DTT). Proteins were eluted from antibody beads in 2.5 ml of HEMGT-150 buffer containing an Rbf2-specific peptide (1 mg/ml) for 2 h at room temperature. One milliliter of the eluted proteins was bound to 100 μl Q Sepharose in a batch for 1 h at room temperature, and bound proteins were eluted with HEMGT buffer containing 650 mM NaCl, separated by 12.5% SDS-PAGE, and stained with the fluorescent dye SYPRO (Sigma, St. Louis, MO). Bands that were enriched with Rbf2-specific antibodies were excised from the gel and analyzed by mass spectrometry.
Gel Filtration Chromatography
Drosophila embryo (0–12 h) extracts (∼2 mg) were fractionated through a Superdex 200 size exclusion column (Amersham, Piscataway, NJ) in HEMGT-100 buffer using an AKTA chromatography system (Amersham). Fractions of 500 μl were collected and alternate fractions were separated by SDS-PAGE and analyzed by Western blotting. Size markers (Sigma MW-GF-1000) were separated under similar conditions.
RNAi and Fluorescence-activated Cell Sorting Analysis
Five hundred-base pair exon sequences corresponding to CSN 1-8 were amplified from Drosophila genomic DNA utilizing divergent T7 tagged primer pairs. PCR products were then transcribed utilizing the MEGAscript kit (Ambion, Austin, TX) for RNAi assays essentially as described (Worby et al., 2001). The lacZ encoding region was amplified from pPelican (Barolo et al., 2000). Primer sequences used for the PCR amplification were obtained from the Drosophila RNAi Screening Center (DRSC; http://flyRNAi.org). S2 cells were incubated with double-stranded RNA (dsRNA) for 5 d and were harvested in Laemmli buffer for protein analyses by Western blotting. Alternatively, 1.6 × 106 S2 cells were treated with csn5 dsRNA, and cells were harvested 8 d later and stained with propidium iodide for fluorescence-activated cell sorting (FACS) analysis.
Chromatin Immunoprecipitation
Chromatin was prepared from 0–12-h-old embryos as described (Cavalli and Paro, 1999), except that embryos were disrupted by sonication using a Branson Sonifier (model 250; Danbury, CT) in lysis buffer containing 50 mM Tris, pH 8.0, 10 mM EDTA, and 1% SDS. Chromatin, 100 μl, was incubated with 1 μl (∼1 μg) of the indicated antibodies for 2 h at room temperature. Samples were processed for sequential chromatin immunoprecipitation (ChIP) essentially as described (Hirsch et al., 2004). ChIP from S2 cells was performed as described (Hirsch et al., 2004).
Antibodies
The α-Rbf1 (226.5) and α-Rbf2 (4.5) antibodies have been described previously (Keller et al., 2005). Rabbit α-CSN4 (232.4) antibodies were generated against the peptide CDYRRKFIEAAQRYNELS, α-Baf53 antibodies were generated against the peptide QEYEEAGKSQVERK, α-PONTIN antibodies were generated against the peptide KRSSKHLSEKNNK, and α-REPTIN antibodies were generated against the peptide NRSSKILKEYQDD. The α-CSN5 antibodies were provided by Daniel Chamovitz (Tel Aviv University), and α-HP1 antibodies by Lori Wallrath (University of Iowa, Iowa City). α-CSN1 antibodies were purchased from BioMol (Plymouth Meeting, PA). The α-cyclin A and α-tubulin antibodies were obtained from Developmental Studies Hybridoma Bank (Iowa).
Rbf Stability Assays
Dechorinated embryos were treated with octane for 3 min and incubated in Schneider medium containing the proteasome inhibitor lactacystin (0, 5, and 10 μM; Sigma) at room temperature for 3 h. Embryos were collected by centrifugation at 5000 rpm for 2 min, washed with PBS, and sonicated in HEMGT-150 buffer containing protease inhibitors and 1 mM DTT. Extracts containing 50 μg of total protein were analyzed by Western blotting. Alternatively, ∼1.4 × 106 S2 cells were incubated for 5 d with 40 μg of dsRNA against CSN4 or lacZ, as a negative control. S2 cells were then treated with dimethyl sulfoxide (DMSO) or DMSO containing 40 μg/ml the proteasome inhibitor MG132 (Sigma) for an additional 6 or 12 h. Extracts containing ∼100 μg of total protein were separated by 10% SDS-PAGE for Western blot analysis.
For stability assays using cycloheximide, ∼4 × 106 S2 cells were treated with cycloheximide (Sigma-Aldrich) at a final concentration of 100 μM for the indicated times. Treated cells were subsequently incubated for 1 h in lysis buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.02% sodium azide, 1% Triton X-100, and Roche Complete Protease inhibitor cocktail; Alameda, CA). After three freeze-thaw cycles the protein concentration in each extract was measured by Bradford assay, and equivalent amounts of total protein for each condition were analyzed by Western blotting for Rbf2 and tubulin. For the experiments examining the effects of Csn5 RNAi and cycloheximide, S2 cells were suspended in serum-free media (1.3 × 106 cells/ml). For each condition, one milliliter of cells was left untreated or was treated with 20–30 μg of Csn5-specific dsRNA for 30 min at 25°C. Three milliliters of complete media was then added to each well. After 5 d, the cells were treated with cycloheximide (100 μM) for times indicated in the figure. Whole cell extracts were prepared, as described above, for Western analysis. Rbf2 levels were quantified using ImageJ software (NIH).
Fly Stocks
The csn4k08018 (Stock number 10765) and csn5L4032 (Stock number 10301) fly stocks were obtained from the Bloomington Stock Center.
RESULTS
Rbf2 Associates with the COP9 Signalosome
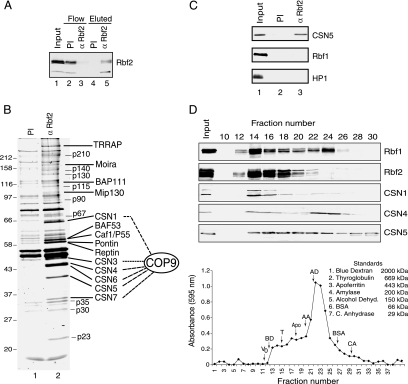
As a first step toward understanding the function of Rbf2 during development, we sought to identity factors associated with Rbf2 during embryogenesis. Rbf2 was purified from embryo nuclear extracts using α-Rbf2 antibodies or with preimmune serum as a negative control, and associated proteins were recovered by competitive elution with the Rbf2-specfic peptide. Endogenous Rbf2 was effectively depleted from the extract with the α-Rbf2 antibody and was recovered by peptide elution from this sample but not from the preimmune antibodies (Figure 1A). To identify proteins associated with Rbf2, those samples recovered by Rbf2-specific peptide elution were separated by SDS-PAGE for subsequent identification by mass spectrometric analysis (Figure 1B). Recovered proteins include components of histone acetyltransferase complexes (TRRAP), ATP-dependent chromatin remodeling complexes (Moira, BAP111, BAF53, Caf1/P55, Pontin, Reptin), dREAM (Mip130), and the COP9 signalosome complex (CSN1, CSN3, CSN4, CSN5, CSN6, CSN7). As shown in Figure 1C, CSN5 was specifically enriched in immunoprecipitation with the α-Rbf2 antibodies and not with the preimmune serum. Consistent with previous findings (Korenjak et al., 2004; Lewis et al., 2004), E2F2 and Caf1/p55 were also enriched (not shown), whereas HP1 and Rbf1 were not associated with Rbf2.
Figure 1.
The COP9 signalosome associates with Rbf2. (A) α-Rbf2 antibodies efficiently deplete Rbf2 from nuclear extracts. αRbf2 Western blot analysis of embryo extracts before (lane 1) and after immunodepletion with preimmune (PI) or α-Rbf2 antibodies (lanes 2 and 3, respectively). Rbf2 was recovered from α-Rbf2 antibodies (lane 5) but not from preimmune antibodies (lane 4) by competitive elution with Rbf2-specific peptide. (B) Eluted proteins were separated by 12.5% SDS PAGE and stained with SYPRO. Those Rbf2-associated proteins whose identity was unambiguously identified are indicated. (C) Rbf2 antibodies specifically precipitate CSN5 from Drosophila embryo extracts. Peptide eluted material from preimmune serum and α-Rbf2 antibodies was analyzed by Western blotting with the antibodies indicated. Specific enrichment of CSN5 but not Rbf1 or HP1 was detected. (D) CSN subunits cofractionate with Rbf1 and Rbf2. α-Rbf1, α-Rbf2, α-CSN1, α-CSN4, and α-CSN5 Western blot analysis was performed for fractions generated by gel filtration chromatography of Drosophila embryo extracts. Total protein levels, as measured by Bradford assay, are indicated by the graph in the bottom panel. Molecular-weight markers were subsequently fractionated under similar conditions, and their relative peak positions are indicated.
Previous studies indicate that RB may regulate target gene expression by modification of local chromatin structure (Harbour and Dean, 2000), and thus the presence of chromatin modifying proteins was not unexpected. The COP9 signalosome, however, has not been directly linked to RB function, and thus its presence in purified Rbf2 fractions was unanticipated. To further examine the connection between Rbf factors and the COP9 signalosome, the association between these factors was analyzed by size exclusion chromatography of Drosophila embryo extracts. As shown in Figure 1D, size fractionation of embryo extracts shows that CSN1, CSN4, and CSN5 copurified with both Rbf1 and Rbf2. CSN4 and CSN5 are also found in smaller complexes or as monomers, as was previously observed (Oron et al., 2002). Together, these data indicate that Rbf2, and possibly Rbf1, can physically associate with the COP9 signalosome complex. Interestingly, the faster migrating hypo-phosphorylated form of Rbf2 (data not shown) cofractioned with COP9 signalosome, whereas the hyper-phosphorylated Rbf2 exhibited a different fractionation pattern distinct from the COP9 signalosome, suggesting that Rbf2 phosphorylation may influence association with the COP9 signalosome.
The COP9 Signalosome Influences Rbf Levels and Cell Cycle Progression
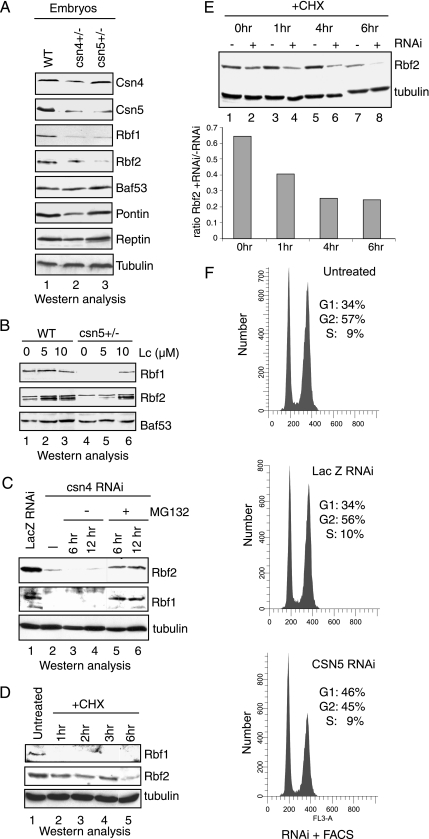
The COP9 signalosome influences the stability of a variety of proteins, including p27, c-Jun, cyclin E, and p53 (Wei and Deng, 2003). To test whether Rbf1 and Rbf2 levels were affected in COP9 mutant backgrounds, embryos were collected from csn4 and csn5 heterozygotes for extract preparation and Western analysis with α-Rbf1 or α-Rbf2 antibodies. As these mutations in csn4 and csn5 are lethal, embryos from homozygotes could not be collected. Embryonic lysates of heterozygous crosses showed that Rbf1 levels were markedly reduced in both csn4 and csn5 embryos, whereas Rbf2 levels were more noticeably reduced in the embryos from the csn5 cross than from the csn4 cross (Figure 2A). No significant changes were consistently noted in levels of other Rbf2-associated proteins, Baf53, Pontin, and Reptin, although Pontin levels are sometimes reduced in the csn4 embryos. Tubulin levels also remained unchanged. Embryonic CSN5 levels were reduced about twofold in both crosses, whereas CSN4 levels were modestly reduced in the csn4 but not csn5 embryos. Together these data suggest that reductions in COP9 signalosome subunit levels can have profound effects on Rbf1 and Rbf2 and indicate that endogenous COP9 subunits stabilize Rbf1 and Rbf2 proteins during development.
Figure 2.
The COP9 signalosome protects Rbf1 and Rbf2 from proteasome-mediated degradation. (A) Reduced levels of Rbf1 and Rbf2 proteins in csn4 and csn5 mutant embryos. Extracts were prepared from 0- to 12-h-old embryos that were collected from wild-type (lane 1), csn4k08018 (lane 2), and csn5L4032 (lane 3) heterozygous flies. Extracts containing 50 μg of total proteins were analyzed by Western blotting to those proteins shown on the right. (B) Inhibition of the proteasome in embryos stabilizes Rbf1 and Rbf2 in a csn5L4032 background. Western analysis was performed using extracts prepared from wild-type embryos (lanes 1–3) or embryos collected from csn5L4032 heterozygous flies (lanes 4–6) and treated for 3 h with of the proteasome inhibitor lactacystin at 0, 5, and 10 μM. (C) Inhibition of the proteasome stabilizes Rbf1 and Rbf2 in S2 cells treated with dsRNA for Csn4. Drosophila S2 cells were incubated with 40 μg of dsRNA specific for lacZ (lane 1) or for csn4 (lanes 2–6). After dsRNA treatment, cells were treated with DMSO or DMSO containing the proteasome inhibitor MG132 (40 μg/ml) for 6 and 12 h as indicated. Lysates were prepared and analyzed by Western blotting for Rbf1 and Rbf2. Equivalent loading of total protein in each lane was verified by anti-tubulin Western analyses. (D) Rbf1 and Rbf2 exhibit different stability after cycloheximide treatment. S2 cells were left untreated (lane 1) or were treated with 100 mM cycloheximide (lanes 2–5) for the indicated times. Rbf1, Rbf2, and tubulin levels were examined by Western blot analyses. (E) Reduced stability of Rbf2 in CSN5-depleted cells. S2 cells were left untreated or were treated with csn5-specific dsRNA for 5 d. Cells were then treated with cycloheximide for the indicated times, and the levels of Rbf2 and tubulin were examined by Western blot analyses. The ratios of Rbf2 in cells for csn5 RNAi plus cycloheximide-treated cells to Rbf2 in cells treated with cycloheximide only are graphed below. Rbf2 levels were quantified using ImageJ (NIH) software. (F) CSN5 knockdown affects G1/S phase progression. S2 cells were incubated with csn5 dsRNA or lacZ dsRNA, as a negative control, and were harvested 8 d after treatment for FACS analyses. CSN5 knockdown resulted in increased number of cells in G1/S phase compared with untreated or lacZ dsRNA-treated cells.
In humans, levels of the RB family member p130 are modulated by cell cycle–dependent ubiquitylation directed by the E3 ubiquitin ligase SCFSkp2 (Tedesco et al., 2002). However, whether any system is engaged to protect RB family members from destruction is unclear. The COP9 complex regulates proteasome-mediated degradation of target proteins, in part by deneddylation of the proteasome-associated E3 cullin subunits (Yang et al., 2002). Thus, the observation that Rbf2 associates with the COP9 signalosome suggests that the absence of the COP9 signalosome in Drosophila may render Rbf proteins more sensitive to destruction by the proteasome. To determine if proteasome activity was indeed required for Rbf instability in COP9 mutants and in knockdown experiments, Rbf1 and Rbf2 levels were assayed in embryos treated with the proteasome inhibitor lactacystin (Figure 2B). Reduced Rbf1 and Rbf2 levels were observed in the csn5 mutant embryos, as expected; however, this effect was reversed by lactacystin treatment, indicating that the destabilization of Rbf proteins associated with diminished COP9 signalosome levels appears to involve proteasome activity. Rbf2 levels were similarly increased in wild-type embryos during lactacystin treatment, suggesting that Rbf2 is naturally sensitive to proteasome activity.
To further explore the role of regulated proteolysis in Rbf metabolism, double-stranded RNA specific for the CSN4 subunit of the COP9 signalosome was incubated with S2 cells in the absence or presence of the proteasome inhibitor MG132, and effect on Rbf1 and Rbf2 was assessed by Western analysis (Figure 2C). Similar to the effect for reduced COP9 signalosome levels in embryos, both Rbf1 and Rbf2 were reduced by the targeted reduction of csn4. Rbf1 and Rbf2 levels were increased in Csn4 dsRNA-treated cells when treated with MG132, indicating protection from proteasome-mediated destruction, whereas this effect was not observed in DMSO-treated cells. Together, these data indicate that Rbf1 and Rbf2 are subject to degradation by a proteasome-dependent pathway, and the COP9 signalosome protects Rbf1 and Rbf2 from such turnover. Interestingly, although Rbf1 and Rbf2 are highly related, these proteins exhibit dramatically different half-lives. In S2 cells treated with cycloheximide (Figure 2D), Rbf1 was unstable with levels becoming undetectable after 1 h. In contrast, Rbf2 levels remained relatively stable until 6 h after treatment. Further measurements after 6 h were unreliable because total protein levels decrease, presumably because of cell death. As shown in Figure 2E, cells treated with both cycloheximide and Csn5-specific dsRNA exhibited accelerated Rbf2 reduction relative to cells treated with cycloheximide alone, consistent with the idea that loss of COP9 signalosome activity decreases the stability of Rbf2.
The loss of Rbf1 and Rbf2 associated with decreased COP9 function would be expected to de-repress Rbf target genes with a concomitant effect on cell cycle progression. Therefore, the effect of CSN5 depletion on cell cycle status after RNAi treatment of S2 cells was examined by FACS analysis. As shown in Figure 2F, CSN5 depletion was associated with a relative increase in the numbers of cells in G1, compared with either untreated S2 cells or cells treated with dsRNA for lacZ. This result is consistent with a recent report identifying the COP9 signalosome as a member of the G1 cell cycle cluster in an RNAi screen for cell cycle progression genes (Bjorklund et al., 2006).
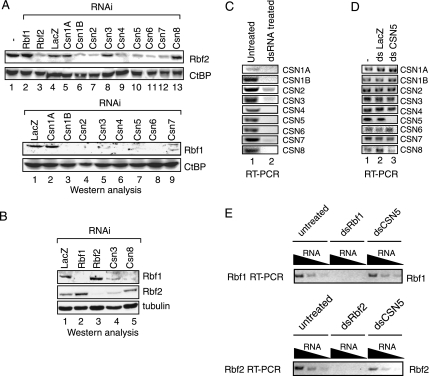
Six COP9 subunits were coimmunoprecipitated with Rbf2, suggesting the entire COP9 complex associates with Rbf proteins; however, significant amounts of CSN4 and CSN5 also fractionated separately from the entire COP9 signalosome. It is thus possible that CSN4 and CSN5, but not the COP9 signalosome, modulate Rbf stability. Therefore, to determine whether all proteins of the COP9 signalosome complex functionally regulate Rbf proteins, as was suggested by the recovery of multiple COP9 subunits during biochemical fractionation, each member of the COP9 complex was individually depleted in cultured S2 cells using RNAi methodology (Figure 3A). As expected, in control treatments, Rbf2 protein levels were selectively reduced by RNAi against Rbf2, whereas Rbf2 protein levels were unaffected by Rbf1-specific RNAi (lanes 2 and 3). dsRNA against lacZ had a reproducible but modest nonspecific effect on Rbf2 protein levels. Strikingly, both Rbf1 and Rbf2 protein levels were sharply reduced in cells treated with dsRNA specific to all COP9 components except CSN1a (Figure 3A and data not shown). Somewhat less robust effects on Rbf2 were obtained with RNAi to CSN3 and CSN8, and as shown in Figure 3B, both Rbf1 and Rbf2 levels can be reduced during Csn3 reduction, whereas Rbf2 is not affected by Csn8 reduction. Thus, the COP9 signalosome may invoke different mechanisms to affect the stability of the different Rbf proteins. As expected, RNAi treatment specific for each COP9 component was observed to strongly reduce levels of the cognate targeted mRNA as measured by RT-PCR reactions (Figure 3C). The specificity of the RNAi treatment is shown by RT-PCR analysis of csn genes: treatment with dsRNA to csn5 selectively reduced only csn5 mRNA, but not mRNAs of other COP9 subunits (Figure 3D). Steady state mRNA levels of rbf1 and rbf2 were largely unaffected by treatment of S2 cells with csn5 dsRNA (Figure 3E), consistent with the data that suggest COP9 signalosome regulation of Rbf1 and Rbf2 functions at the level of protein turnover. Together, these results show that the COP9 signalosome plays a positive role in maintaining Rbf1 and Rbf2 levels, probably through a posttranscriptional mechanism.
Figure 3.
Multiple COP9 subunits participate in Rbf1 and Rbf2 stabilization. (A) Rbf1 and Rbf2 protein levels are reduced after treatment of cultured S2 cells with dsRNA for COP9 signalosome subunits. Lysates were prepared and analyzed by Western blotting for Rbf1, Rbf2, and for CtBP, as a negative control. (B) Rbf1 and Rbf2 are differentially sensitive to CSN8 knockdown. S2 cells were treated with the indicated dsRNAs followed by Western blot analyses of Rbf1, Rbf2, and tubulin. Rbf1 but not Rbf2 levels were decreased in response to csn8-specific RNAi treatment. (C) Effect of RNAi on cognate mRNAs. Double-stranded RNA specific for each COP9 subunit effectively reduces its cognate target RNA in S2 cells, as measured by RT-PCR. (D) Specificity of RNAi. dsRNA specific for csn5 does not affect steady state mRNA levels for the other COP9 subunits, as measured by RT-PCR. (E) csn5-specific dsRNA does not substantively affect rbf1 or rbf2 mRNA, as measured by RT-PCR. Double-stranded rbf1 and rbf2 effectively deplete the cognate mRNAs, as expected.
Simultaneous Promoter Association by CSN4 and RB Family Members
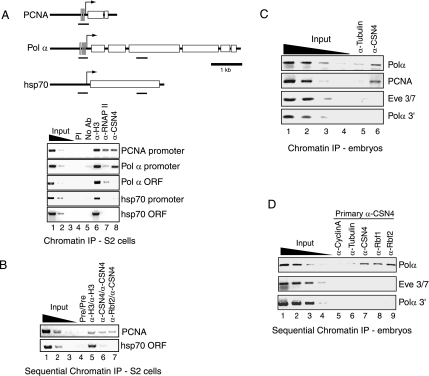
The previous experiments do not indicate whether the Rbf–COP9 interaction takes place on or off DNA; thus we tested directly whether the COP9 signalosome might be promoter associated. Rbf1 and Rbf2 were previously shown to associate directly with promoters of target genes (Stevaux et al., 2002, 2005), and as expected, both Polα and PCNA promoter regions were enriched in α-Rbf1 and α-Rbf2 ChIP reactions from Drosophila embryos (data not shown). Next, we tested whether the COP9 signalosome could also associate with Rbf-targeted promoters in S2 cells. As shown in Figure 4A, the Polα and PCNA promoter regions were enriched in α-CSN4 antibody immunoprecipitation reactions, whereas the open reading frame (ORF) regions of Polα or hsp70 were not. No enrichment was observed with preimmune serum. RNA polymerase II was detected at the PCNA promoter, but not at the Polα or hsp70 regions, whereas all loci examined were enriched using α-histone H3 antibodies. Therefore, CSN4 and potentially the COP9 signalosome complex localize to Rbf target genes. Enrichment of promoter regions of other developmentally regulated genes unrelated to Rbf pathways, including zen, ftz, and tll, were also observed during CSN4 immunoprecipitation (data not shown), suggesting that CSN4 and potentially the COP9 signalosome may play a more general role in transcription.
Figure 4.
CSN4 co-occupies target gene promoters with Rbf1 and Rbf2. (A) CSN4 associates with Rbf target genes in Drosophila S2 cells. Formaldehyde cross-linked chromatin was prepared from Drosophila S2 cells and immunoprecipitated using the indicated antibodies in single-round reactions. Enrichment of the Rbf-regulated Polα and PCNA promoters was observed by α-CSN4 immunoprecipitation reactions (lane 8), but not in reactions lacking antibody (lane 4) or using preimmune serum (PI, lane 5). Lanes 1–3 show the amplification signals for 1, 0.1, and 0.01% of the input chromatin. A schematic of the genes examined is shown at the top with the positions of E2F-binding sites indicated by gray vertical bars. The various regions amplified by PCR are indicated by the small black horizontal bars. (B) Rbf2 and CSN4 co-occupy the PCNA promoter in Drosophila S2 cells. Sequential chromatin immunoprecipitation of S2 cell chromatin was performed with preimmune serum (lane 4), α-histone H3 (lane 5), α-CSN4 (lane 6), or α-Rbf2 antibodies (lane 7). Chromatin was recovered by elution with DTT and SDS followed by dilution for the second-round immunoprecipitation using the indicated antibodies. (C) CSN4 associates with Rbf target genes in Drosophila embryos. Formaldehyde cross-linked chromatin was prepared from 0- to 12-h-old wild-type Drosophila embryos for immunoprecipitation using α-tubulin (lane 5) or α-CSN4 antibodies (lane 6). Lanes 1–4 show the amplification signal for 10, 1, 0.1, and 0.01% of the input chromatin. (D) CSN4 simultaneously occupies the Polα promoter along with Rbf1 and Rbf2 in Drosophila embryos. Primary immunoprecipitation using formaldehyde cross-linked chromatin from 0- to 12-h-old embryos was carried out first with α-CSN4 antibody followed by a second immunoprecipitation with the indicated antibodies. Enrichment of the Polα but not the eve stripe 3/7 enhancer or Polα 3′ region was observed in the second-round Rbf1 and Rbf2 immunoprecipitation reactions.
Sequential ChIP experiments were then performed to determine whether Rbf proteins and CSN4 simultaneously occupy the PCNA promoter (Figure 4B). Indeed, the PCNA promoter but not the hsp70 ORF was enriched in α-Rbf2/α-CSN4 double IP, at levels comparable to that observed for the α-CSN4/α-CSN4 double IP or α-histone H3/α-histone H3 double IP. Similar experiments were performed to determine whether CSN4 could associate with target gene promoters in the developing embryo (Figure 4C). Again, both Polα and PCNA promoter regions were specifically enriched in α-CSN4 ChIP reactions. Significant enrichment of the Polα 3′ gene region or even-skipped (eve) stripe 3/7 enhancer region was not observed. Sequential ChIP experiments from embryos also revealed significant enrichment of the Polα promoter, but not the Polα 3′ region or eve stripe 3/7 enhancer in reactions performed first using α-CSN4 antibodies followed by immunoprecipitation using α-CSN4, α-Rbf1, or α-Rbf2 antibodies. In contrast, reactions performed using either α-cyclin A or α-tubulin antibodies in the second round showed no significant enrichment for these regions (Figure 4D). We conclude that Rbf1 and Rbf2 can simultaneously occupy the Polα promoter along with CSN4.
DISCUSSION
Retinoblastoma family proteins are important regulators of cell proliferation and differentiation through the regulation of critical genes that control these processes. The ability of Retinoblastoma family proteins to control gene expression at defined times in specific tissues depends on a wide variety of coregulatory factors that enact RB repression in gene-specific manner. Herein, we provide evidence supporting a novel link between Drosophila Rbf2 and the COP9 signalosome, an evolutionarily conserved complex that is essential for proper developmental patterns from plants to mammals.
The role of the COP9 signalosome in gene regulation by Rbf proteins remains imprecisely defined; however, our data suggest that the COP9 signalosome protects Rbf1 and Rbf2 from proteasome-mediated destruction. Rbf protein levels were reduced in csn4 and csn5 mutant embryos, and embryonic levels of both Rbf proteins were restored by inhibiting the proteasome. Similarly, the destruction of Rbf1 and Rbf2 in S2 cells treated with csn4-specific dsRNA was similarly blocked by inhibition of the proteasome. Furthermore, RNAi-mediated reduction of multiple COP9 signalosome subunits lead to reduced Rbf1 and Rbf2 levels, indicating that the entire COP9 signalosome complex is involved in this function. The observed protection of Rbf1 and Rbf2 may extend from two aspects of the COP9 signalosome. First, many subunits of the COP9 signalosome share limited sequence homology with components of the 19S proteasome lid complex (Wei et al., 1998; Henke et al., 1999), and thus the COP9 signalosome may compete with the proteasome for access to Rbf proteins. Second, the COP9 signalosome can deneddylate the cullin subunits of SCF ubiquitin E3 ligase complexes (Lyapina et al., 2001); therefore, altered SCF complex activity in the absence of the COP9 signalosome may be directly responsible for downstream changes in Rbf1 and Rbf2 levels. If so, the decreased levels of Rbf1 and Rbf2 as seen in Drosophila embryos, possibly via a SCF ubiquitin E3 ligase pathway, would be similar to the SCF-mediated destruction of p130, observed in humans (Tedesco et al., 2002). However, SCF deneddylation appears to play both positive and negative roles for SCF activity and subsequent target protein destruction depending on species and cell type examined (Wu et al., 2006), and thus, the COP9 signalosome may similarly exhibit bipolar effects on Rbf1 and Rbf2 protection, depending on context. At least in early stages of Drosophila development, the COP9 signalosome plays a protection role in Rbf function.
Previous studies have implicated the COP9 signalosome complex in cell cycle regulatory pathways during development, and individually, the mammalian CSN5 protein, also known as Jab1, has recently been shown to bind E2F1 (Hallstrom and Nevins, 2006), a protein partner for Rbf1. The newly described linkage between the Drosophila Retinoblastoma protein Rbf2 and COP9 signalosome is consistent with a role for COP9 signalosome in cell cycle progression through its association with Rbf proteins. However, depletion of CSN5 by RNAi resulted in blocked G1/S progression, whereas loss of Rbf1 and Rbf2 function would be expected to facilitate G1/S progression. Thus, in our experiments, it appears likely that impaired function of other cell cycle regulatory proteins such as E2F and cyclin E in the absence of COP9 signalosome activity may play a dominant role in limiting cell cycle progression through G1 phase.
The COP9 signalosome has also been suggested to play an important role in modulating cancer initiation and progression (Richardson and Zundel, 2005). In this arena, a number of factors that play critical roles in cellular proliferation, including the cyclin/cdk inhibitor p27, cyclin E, c-jun, and the tumor suppressor p53, among others, have been previously linked to the COP9 signalosome. Thus, one mechanism for the tumorigenic control by COP9 is through its targeting of proto-oncoproteins and tumor suppressor proteins that play critical roles in governing cellular proliferation. Our data linking the COP9 signalosome to Rbf proteins, homologues of the human Retinoblastoma tumor suppressor protein, strengthens this connection. Interestingly, the CSN4 subunit of the COP9 signalosome co-occupies selected target gene promoters along with Rbf2. The presence of this COP9 complex subunit at Rbf1 and Rbf2 target gene promoters indicates that the complex may play a direct role in transcriptional regulation, or alternatively, the COP9 signalosome may stabilize Rbf proteins against degradation because these proteins regulate gene expression during growth. Interestingly, the presence of proteasome subunits has been documented at actively transcribed genes (Gonzalez et al., 2002), and ubiquitylation and proteasome-mediated destruction of transcriptional activators has been linked to increased activator potency (Salghetti et al., 2000; Lipford et al., 2005). As activator ubiquitylation can serve as a marker for coregulatory protein recruitment (Kurosu and Peterlin, 2004), it will be important to determine whether repressor potency and corepressor recruitment are likewise linked to signals that govern their own destruction.
ACKNOWLEDGMENTS
We thank Louis King and the MSU Flow Cytometry Center, as well as the MSU proteomics facility, for invaluable technical assistance. We thank Nick Dyson, Lori Wallrath, and Daniel Chamovitz for sharing antibodies and Carla Margulies for preparation of Drosophila nuclear extracts. This work was supported by National Institutes of Health Grant R01 GM59805 to R.W.H. and a grant from the MSU foundation to D.N.A. and R.W.H.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0790) on January 24, 2007.
REFERENCES
- Barolo S., Carver L. A., Posakony J. W. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29:726–732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- Beall E. L., Manak J. R., Zhou S., Bell M., Lipsick J. S., Botchan M. R. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature. 2002;420:833–837. doi: 10.1038/nature01228. [DOI] [PubMed] [Google Scholar]
- Bjorklund M., Taipale M., Varjosalo M., Saharinen J., Lahdenpera J., Taipale J. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature. 2006;439:1009–1013. doi: 10.1038/nature04469. [DOI] [PubMed] [Google Scholar]
- Cavalli G., Paro R. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science. 1999;286:955–958. doi: 10.1126/science.286.5441.955. [DOI] [PubMed] [Google Scholar]
- Classon M., Dyson N. p107 and p130, versatile proteins with interesting pockets. Exp. Cell Res. 2001;264:135–147. doi: 10.1006/excr.2000.5135. [DOI] [PubMed] [Google Scholar]
- Classon M., Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- Dimova D. K., Stevaux O., Frolov M. V., Dyson N. J. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 2003;17:2308–2320. doi: 10.1101/gad.1116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronkin S., Djagaeva I., Beckendorf S. K. The COP9 signalosome promotes degradation of Cyclin E during early Drosophila oogenesis. Dev. Cell. 2003;4:699–710. doi: 10.1016/s1534-5807(03)00121-7. [DOI] [PubMed] [Google Scholar]
- Frolov M. V., Dyson N. J. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J. Cell Sci. 2004;117:2173–2181. doi: 10.1242/jcs.01227. [DOI] [PubMed] [Google Scholar]
- Gonzalez F., Delahodde A., Kodadek T., Johnston S. A. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science. 2002;296:548–550. doi: 10.1126/science.1069490. [DOI] [PubMed] [Google Scholar]
- Hallstrom T. C., Nevins J. R. Jab1 is a specificity factor for E2F1-induced apoptosis. Genes Dev. 2006;20:613–623. doi: 10.1101/gad.1345006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour J. W., Dean D. C. Chromatin remodeling and Rb activity. Curr. Opin. Cell Biol. 2000;12:685–689. doi: 10.1016/s0955-0674(00)00152-6. [DOI] [PubMed] [Google Scholar]
- Henke W., Ferrell K., Bech-Otschir D., Seeger M., Schade R., Jungblut P., Naumann M., Dubiel W. Comparison of human COP9 signalosome and 26S proteasome ‘lid'. Mol. Biol. Rep. 1999;26:29–34. doi: 10.1023/a:1006991419464. [DOI] [PubMed] [Google Scholar]
- Hirsch H. A., Gu L., Henry R. W. The retinoblastoma tumor suppressor protein targets distinct general transcription factors to regulate RNA polymerase III gene expression. Mol. Cell. Biol. 2000;20:9182–9191. doi: 10.1128/mcb.20.24.9182-9191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch H. A., Jawdekar G. W., Lee K. A., Gu L., Henry R. W. Distinct mechanisms for repression of RNA polymerase III transcription by the retinoblastoma tumor suppressor protein. Mol. Cell. Biol. 2004;24:5989–5999. doi: 10.1128/MCB.24.13.5989-5999.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller S. A., Ullah Z., Buckley M.S., Henry R. W., Arnosti D. N. Distinct developmental expression of Drosophila retinoblastoma factors. Gene Expr. Patterns. 2005;5:411–421. doi: 10.1016/j.modgep.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Korenjak M., Taylor-Harding B., Binne U. K., Satterlee J. S., Stevaux O., Aasland R., White-Cooper H., Dyson N., Brehm A. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119:181–193. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Kurosu T., Peterlin B. M. VP16 and ubiquitin; binding of P-TEFb via its activation domain and ubiquitin facilitates elongation of transcription of target genes. Curr. Biol. 2004;14:1112–1116. doi: 10.1016/j.cub.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Lewis P. W., Beall E. L., Fleischer T. C., Georlette D., Link A. J., Botchan M. R. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 2004;18:2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford J. R., Smith G. T., Chi Y., Deshaies R. J. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–116. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- Lyapina S., Cope G., Shevchenko A., Serino G., Tsuge T., Zhou C., Wolf D. A., Wei N., Deshaies R. J. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- Morris E. J., Dyson N. J. Retinoblastoma protein partners. Adv. Cancer Res. 2001;82:1–54. doi: 10.1016/s0065-230x(01)82001-7. [DOI] [PubMed] [Google Scholar]
- Mulligan G., Jacks T. The retinoblastoma gene family: cousins with overlapping interests. Trends Genet. 1998;14:223–229. doi: 10.1016/s0168-9525(98)01470-x. [DOI] [PubMed] [Google Scholar]
- Nevins J. R. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- Oron E., Mannervik M., Rencus S., Harari-Steinberg O., Neuman-Silberberg S., Segal D., Chamovitz D. A. COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development. 2002;129:4399–4409. doi: 10.1242/dev.129.19.4399. [DOI] [PubMed] [Google Scholar]
- Paggi M. G., Baldi A., Bonetto F., Giordano A. Retinoblastoma protein family in cell cycle and cancer: a review. J. Cell Biochem. 1996;62:418–430. doi: 10.1002/(SICI)1097-4644(199609)62:3%3C418::AID-JCB12%3E3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Richardson K. S., Zundel W. The emerging role of the COP9 signalosome in cancer. Mol. Cancer Res. 2005;3:645–653. doi: 10.1158/1541-7786.MCR-05-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salghetti S. E., Muratani M., Wijnen H., Futcher B., Tansey W. P. Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl. Acad. Sci. USA. 2000;97:3118–3123. doi: 10.1073/pnas.050007597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers W. R., Kaelin W. G., Jr Role of the retinoblastoma protein in the pathogenesis of human cancer. J. Clin. Oncol. 1997;15:3301–3312. doi: 10.1200/JCO.1997.15.11.3301. [DOI] [PubMed] [Google Scholar]
- Stevaux O., Dimova D., Frolov M. V., Taylor-Harding B., Morris E., Dyson N. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J. 2002;21:4927–4937. doi: 10.1093/emboj/cdf501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevaux O., Dimova D. K., Ji J. Y., Moon N. S., Frolov M. V., Dyson N. J. Retinoblastoma family 2 is required in vivo for the tissue-specific repression of dE2F2 target genes. Cell Cycle. 2005;4:1272–1280. doi: 10.4161/cc.4.9.1982. [DOI] [PubMed] [Google Scholar]
- Tedesco D., Lukas J., Reed S. I. The pRb-related protein p130 is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(Skp2) Genes Dev. 2002;16:2946–2957. doi: 10.1101/gad.1011202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N., Deng X. W. COP9, a new genetic locus involved in light-regulated development and gene expression in arabidopsis. Plant Cell. 1992;4:1507–1518. doi: 10.1105/tpc.4.12.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N., Deng X. W. The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 2003;19:261–286. doi: 10.1146/annurev.cellbio.19.111301.112449. [DOI] [PubMed] [Google Scholar]
- Wei N., Tsuge T., Serino G., Dohmae N., Takio K., Matsui M., Deng X. W. The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. Curr. Biol. 1998;8:919–922. doi: 10.1016/s0960-9822(07)00372-7. [DOI] [PubMed] [Google Scholar]
- Worby C. A., Simonson-Leff N., Dixon J. E. RNA interference of gene expression (RNAi) in cultured Drosophila cells. Sci. STKE. 2001;2001:PL1. doi: 10.1126/stke.2001.95.pl1. [DOI] [PubMed] [Google Scholar]
- Wu J. T., Chan Y. R., Chien C. T. Protection of cullin-RING E3 ligases by CSN-UBP12. Trends Cell Biol. 2006;16:362–369. doi: 10.1016/j.tcb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Yang X., Menon S., Lykke-Andersen K., Tsuge T., Di X., Wang X., Rodriguez-Suarez R. J., Zhang H., Wei N. The COP9 signalosome inhibits p27(kip1) degradation and impedes G1-S phase progression via deneddylation of SCF Cul1. Curr. Biol. 2002;12:667–672. doi: 10.1016/s0960-9822(02)00791-1. [DOI] [PubMed] [Google Scholar]