Figure 8.
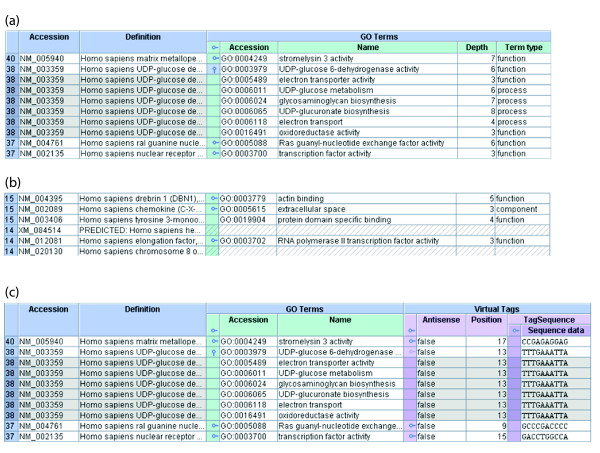
Displaying multiple annotations through expansion points. (a) A detail from the Explorer showing an open expansion point. The blue outer columns represent a set of Refseq genes, the green inner columns represent the GO terms mapped to those genes. As there are many GO annotations per gene, the property is represented by an expansion point. The second expansion point is expanded and the others collapsed. One can see that the expansion is represented by the product of the subject and objects of that property. The repeated subject cells are shaded for clarity. Expansion points in the header allow each property to be expanded or collapsed in bulk per column. (b) A detail from the Explorer showing various expansion point states. Notice that the third Refseq gene does not have an expansion point because it has only one GO annotation. The fourth and sixth genes have no GO annotations at all, as represented by the hatched cells. The first, second and fifth genes have multiple GO annotations, all collapsed. (c) A detail from the Explorer showing conflicting expansion points. The user is now viewing the set of Refseq genes, their GO annotations and their virtual tags (lilac), along with the sequences of those virtual tags (purple). Properties 'GO Terms' and 'Virtual Tags' are sibling one-to-many properties of the class Refseq gene and are, therefore, 'in competition'. The user has expanded the GO annotations of the second Refseq gene and the product is being displayed. Notice that the respective 'expansion point' for the virtual tags property is now dimmed and the first virtual tag is now repeated as part of the GO product. If the dimmed 'expansion point' is expanded, then the open competing property (GO Terms) will automatically be closed.
