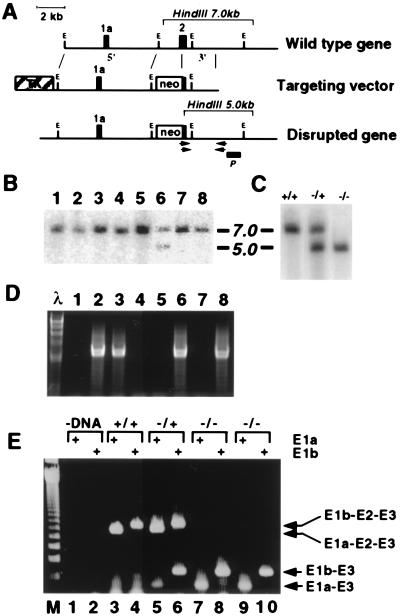
Figure 1.
Disruption of the murine Nfia gene and demonstration of “exon-skipping.” (A) The structure of a portion of the wild-type Nfia gene is shown with exons 1a and 2 as black boxes, EcoRI sites marked E, and the 7.0-kb HindIII fragment shown with a bracket. The targeting vector was constructed as described in Materials and Methods, and recombination within the 5′ and 3′ regions with the wild-type gene would yield the disrupted Nfia gene missing most of exon 2. (B) Clones of ES cells electroporated with the targeting vector were lysed and DNA was isolated, digested with HindIII, transferred to membranes, and probed with the probe shown as P in A. This probe lies outside of the targeting vector and detects a 7.0-kb band from the wild-type Nfia allele and a 5.0-kb band from the disrupted allele. Lanes 1–5, 7, and 8 show a wild-type pattern, while lane 6 shows both the wild-type and disrupted alleles. (C) Genomic DNA from the tails of +/+, +/−, and −/− animals was digested with HindIII and analyzed as in B. (D) The nested primers shown as arrows below the disrupted gene in A were used to amplify genomic DNA of the progeny of a mouse heterozygous for the disrupted Nfia− allele. Lanes 2, 3, 6, and 8 are positive for the 2.3-kb PCR fragment, while lanes 1, 4, 5, and 7 are negative. Size markers of HindIII-digested λ DNA are shown in the leftmost lane, marked λ. The rightmost set of nested primers from A lies outside of the targeting construct. (E) RNA from the brains of wild-type (lanes 3 and 4), heterozygous (+/−, lanes 5 and 6), and homozygous Nfia− mice (−/−, lanes 7–10) was isolated, reverse transcribed, and PCR amplified by using primers in exon 3 and either exon 1a or 1b of Nfia as denoted above the lanes. The products were analyzed on a 2% agarose gel along with 123-bp ladder markers (lane M). The arrows E1b-E2-E3 and E1a-E2-E3 show the positions of correctly spliced NFI-A mRNAs, while the arrows E1b-E3 and E1a-E3 show the positions of the products lacking exon 2. Lanes 1 and 2 contained no reverse transcribed product.