Abstract
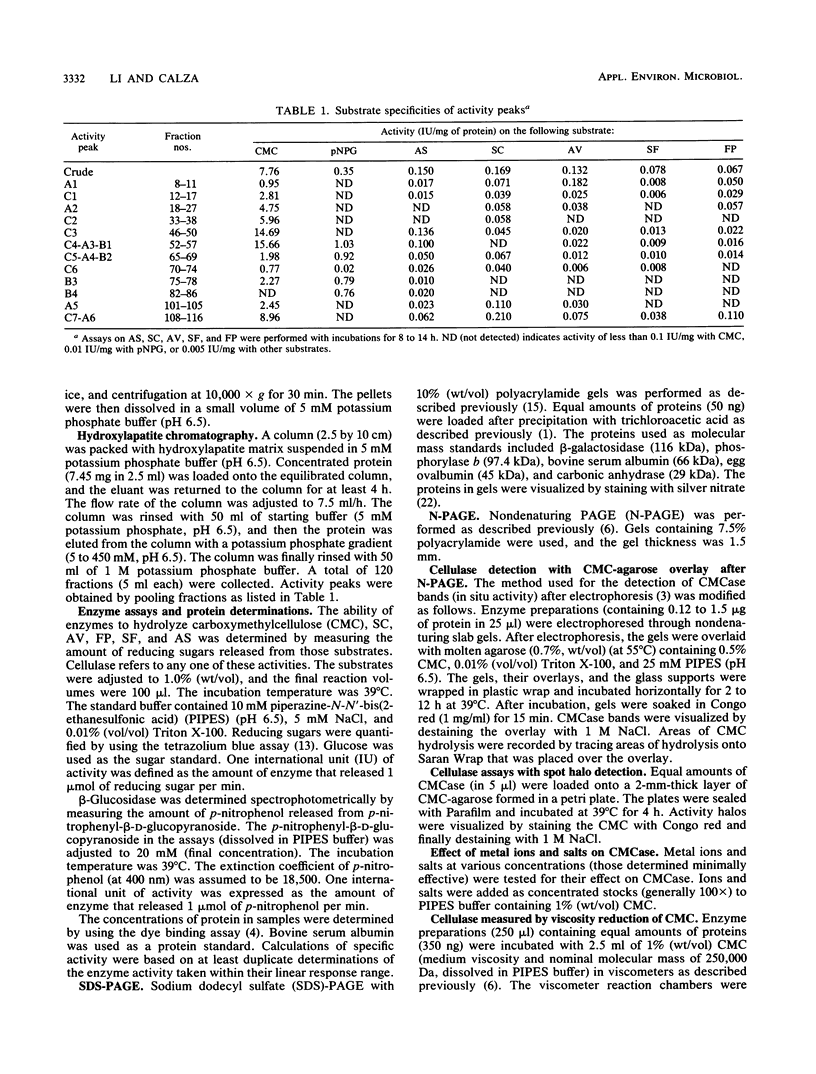
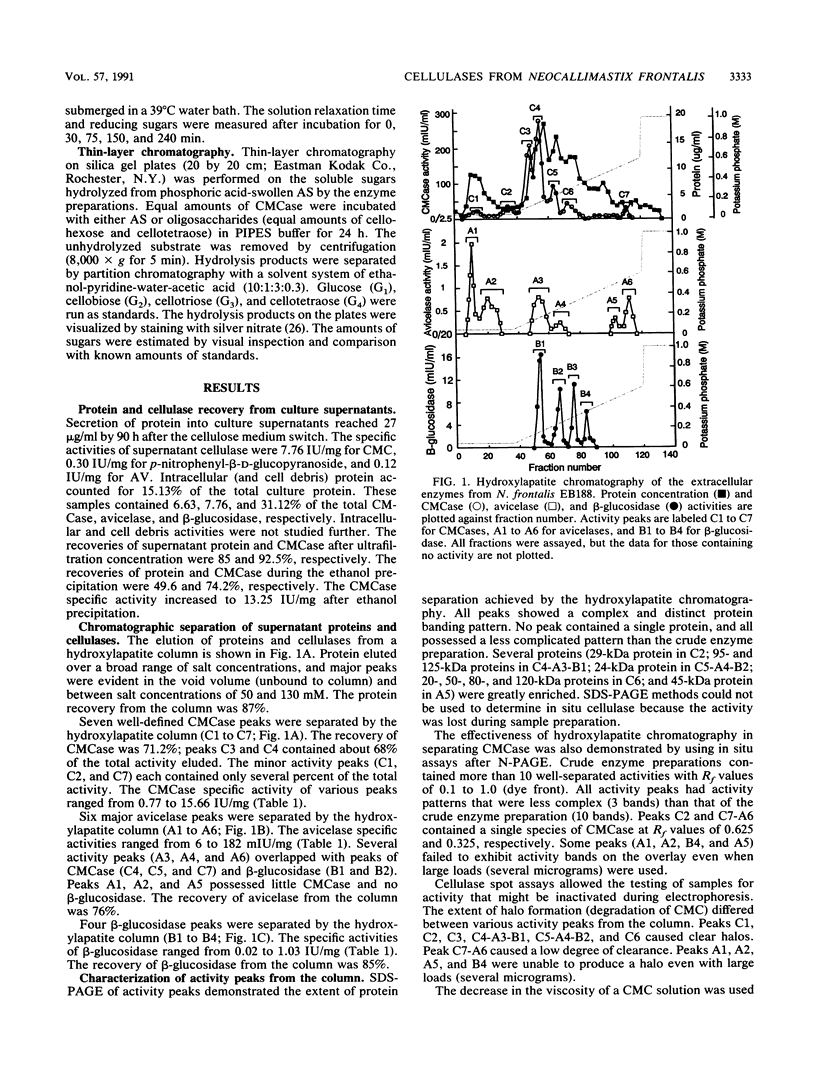
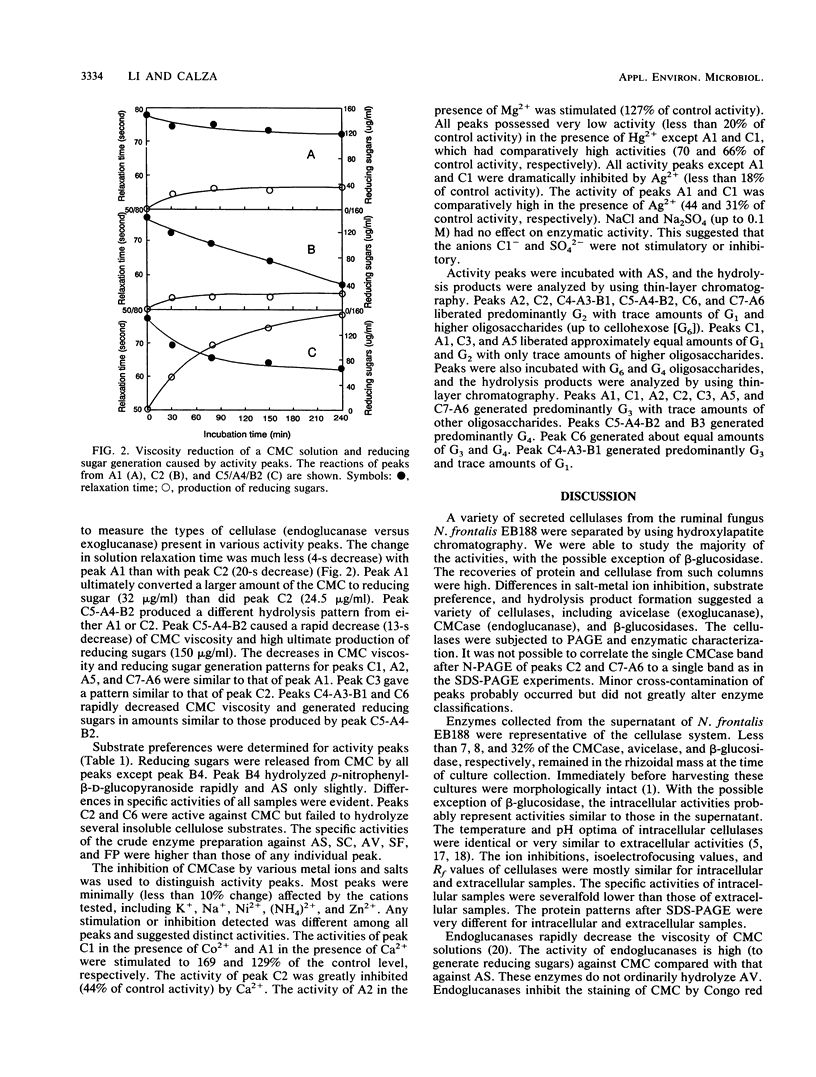
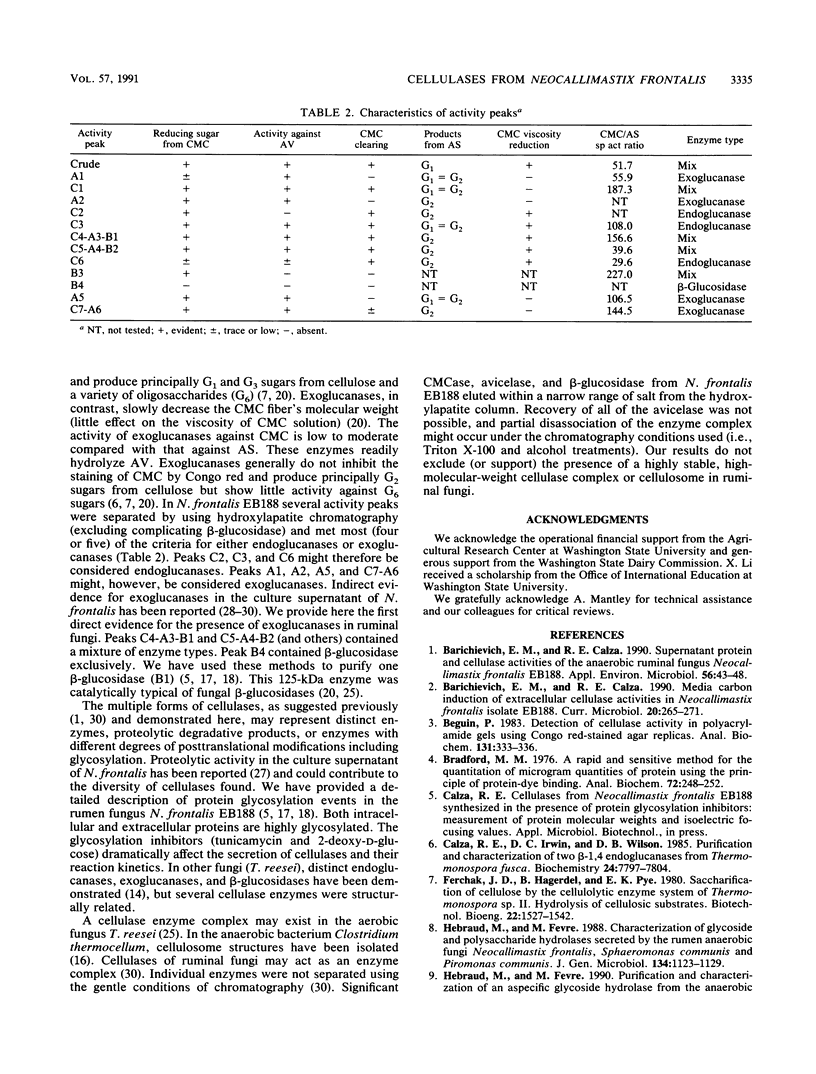
Cellulases from the ruminal fungus Neocallimastix frontalis EB188 were separated by using hydroxylapatite column chromatography. Seven carboxymethylcellulases, six avicelases, and four beta-glucosidases accounted for the majority of the activities. The separation of enzymes was confirmed by using polyacrylamide gel electrophoresis. Electrophoretic migration, analysis of hydrolysis products, and substrate specificity measurements suggested that several different cellulases were secreted in N. frontalis EB188. The possible relationship of cellulase diversity to protein glycosylation is discussed.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barichievich E. M., Calza R. E. Supernatant protein and cellulase activities of the anaerobic ruminal fungus Neocallimastix frontalis EB188. Appl Environ Microbiol. 1990 Jan;56(1):43–48. doi: 10.1128/aem.56.1.43-48.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Béguin P. Detection of cellulase activity in polyacrylamide gels using Congo red-stained agar replicas. Anal Biochem. 1983 Jun;131(2):333–336. doi: 10.1016/0003-2697(83)90178-1. [DOI] [PubMed] [Google Scholar]
- Joblin K. N. Isolation, enumeration, and maintenance of rumen anaerobic fungi in roll tubes. Appl Environ Microbiol. 1981 Dec;42(6):1119–1122. doi: 10.1128/aem.42.6.1119-1122.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jue C. K., Lipke P. N. Determination of reducing sugars in the nanomole range with tetrazolium blue. J Biochem Biophys Methods. 1985 Aug;11(2-3):109–115. doi: 10.1016/0165-022x(85)90046-6. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamed R., Setter E., Bayer E. A. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J Bacteriol. 1983 Nov;156(2):828–836. doi: 10.1128/jb.156.2.828-836.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe S. E., Theodorou M. K., Trinci A. P. Cellulases and xylanase of an anaerobic rumen fungus grown on wheat straw, wheat straw holocellulose, cellulose, and xylan. Appl Environ Microbiol. 1987 Jun;53(6):1216–1223. doi: 10.1128/aem.53.6.1216-1223.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandels M., Andreotti R., Roche C. Measurement of saccharifying cellulase. Biotechnol Bioeng Symp. 1976;(6):21–33. [PubMed] [Google Scholar]
- Mountfort D. O., Asher R. A. Production and regulation of cellulase by two strains of the rumen anaerobic fungus Neocallimastix frontalis. Appl Environ Microbiol. 1985 May;49(5):1314–1322. doi: 10.1128/aem.49.5.1314-1322.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley B. R., Kirsch D. R., Morris N. R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem. 1980 Jul 1;105(2):361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- Pearce P. D., Bauchop T. Glycosidases of the rumen anaerobic fungus Neocallimastix frontalis grown on cellulosic substrates. Appl Environ Microbiol. 1985 May;49(5):1265–1269. doi: 10.1128/aem.49.5.1265-1269.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A. G., Orpin C. G. Glycoside hydrolase enzymes present in the zoospore and vegetative growth stages of the rumen fungi Neocallimastix patriciarum, Piromonas communis, and an unidentified isolate, grown on a range of carbohydrates. Can J Microbiol. 1987 May;33(5):427–434. doi: 10.1139/m87-072. [DOI] [PubMed] [Google Scholar]
- Williams A. G., Orpin C. G. Polysaccharide-degrading enzymes formed by three species of anaerobic rumen fungi grown on a range of carbohydrate substrates. Can J Microbiol. 1987 May;33(5):418–426. doi: 10.1139/m87-071. [DOI] [PubMed] [Google Scholar]


