Abstract
Purpose
The proapoptotic receptors tumor necrosis factor – related apoptosis-inducing ligand receptor 1 (TRAIL-R1) and TRAIL-R2 are targets of drugs in clinical development, and receptor expression levels may be important determinants of sensitivity to receptor agonists. We assessed TRAIL-R1 and TRAIL-R2 expression patterns in a large cohort of melanomas and benign nevi.
Experimental Design
We analyzed tissue microarrays containing 546 melanomas and 540 nevi using our automated quantitative method to measure protein levels in situ (AQUA).The system uses S100 to define pixels as melanoma (tumor mask) within the array spot and measures intensity of TRAIL-receptor expression using Cy5-conjugated antibodies within the mask. AQUA scores were correlated with clinical and pathologic variables.
Results
TRAIL-R1 and TRAIL-R2 expression was higher in melanomas than in nevi (P < 0.0001), and higher in primary than in metastatic specimens (P = 0.0031and P < 0.0001, respectively). TRAIL-R1 and TRAIL-R2 expression exceeding the 95th percentile for nevi was found in19% and 74% of melanoma specimens, respectively. Although on univariate analysis, high TRAIL-R2 expression correlated with increased survival (P = 0.0439), it was not associated with survival within the primary or metastatic subcohorts. TRAIL-R1expression was not associated with survival.
Conclusions
TRAIL-R1 and TRAIL-R2 expressionis higher in malignant melanocytes thanin their benign counterparts, suggesting that these receptors might be effective therapeutic targets in melanoma. Expression is higher in early-stage disease than in metastatic specimens, and expression exceeding that found in nevi is found in a substantially larger fraction of melanomas for TRAIL-R2 compared with TRAIL-R1. Assessment of baseline tumor TRAIL receptor expression may be important in analysis of clinical trials involving TRAIL receptor agonists.
The incidence of primary cutaneous melanoma in the United States is increasing faster than that of any other malignancy (1). For patients who develop metastatic disease, the prognosis is poor, with median survival in the range of 9 to 12 months. Although several chemotherapeutic and biological agents have low-level activity in metastatic melanoma, neither single agent nor combination therapy has improved overall survival when compared with observation (2–4).
Various agents targeting specific cell-surface receptors or signaling pathways are in clinical development and are predicted to selectively kill or inhibit malignant cells, thus increasing efficacy while reducing toxicity and offering the possibility of improved therapy for patients with melanoma. Among the most promising therapeutic targets are the tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) cell-surface receptors 1 and 2 (TRAIL-R1 and TRAIL-R2; refs. 5–7). TRAIL-R1 and TRAIL-R2, also known as DR4 and DR5, are members of the tumor necrosis factor receptor gene family. They contain an extracellular domain and a cytoplasmic sequence termed the “death domain,” which the receptors use to engage the apoptotic machinery of the cell (8). Whereas TRAIL-R1 and TRAIL-R2 both seem to contribute to TRAIL-induced apoptosis, their relative contribution in different diseases is unknown. Some studies support a relatively larger contribution of TRAIL-R2 (9) in melanoma whereas others support a larger contribution of TRAIL-R1 (10).
Currently, several agents are in clinical development that specifically target TRAIL-R1 and TRAIL-R2. The TRAIL ligand showed antitumor activity with limited toxicity to normal cells in mouse models (7, 11) and, in combination with chemotherapeutic drugs, produced synergistic antitumor effects against various malignancies in vitro and in vivo (12–18). TRAIL ligand is under investigation in early-phase clinical trials.6 Because of the potential competition between proapoptotic TRAIL receptors and the presence of TRAIL-binding decoy receptors, antibodies that selectively activate the proapoptotic receptors may have therapeutic advantages compared with the TRAIL ligand. Activating monoclonal antibodies to TRAIL-R1 and TRAIL-R2 have shown single agent antitumor activity, as well as synergism with a number of chemotherapeutic agents in vitro (5, 6, 19, 20). These activating antibodies also induced apoptosis in cancer cells in vivo (5, 6). Agonistic monoclonal antibodies to TRAIL-R1 and TRAIL-R2 recently entered clinical development for patients with advanced malignancies.7
A number of published studies support exploration of TRAIL receptor agonists for the treatment of patients with melanoma and also show the potential importance of receptor expression level in controlling tumor cell response to these agents. TRAIL expressed on the surface of activated (but not resting) CD4 T cells induced apoptosis in melanoma cell lines (21) and inhibition of TRAIL decreased CD4 T-cell–mediated cytotoxicity (22). Griffith et al. (23) showed that TRAIL induced apoptosis in two thirds of melanoma cell lines, which were resistant to other tumor necrosis factor family cytokines. In the latter studies, sensitivity to TRAIL-induced apoptosis was correlated to TRAIL-R1 and especially TRAIL-R2 expression (23). Zhang et al. (24) similarly found that TRAIL-R1 and TRAIL-R2 expression was necessary for TRAIL-induced apoptosis of melanoma cell lines. Nguyen et al. (25) confirmed that low TRAIL-R1 and TRAIL-R2 expression was associated with resistance to TRAIL-induced apoptosis in a small number (eight) of fresh melanoma isolates.
Because of the potential importance of receptor expression in determining sensitivity to TRAIL receptor–targeted therapeutics, we assessed the expression of both TRAIL-R1 and TRAIL-R2 in a large cohort of human melanoma specimens and benign nevi, and correlated expression to survival and other prognostic variables. To the best of our knowledge, this is the first large study looking at expression of TRAIL receptors in melanoma specimens. To obtain quantitative, objective measures of expression, we used our newly developed method of automated quantitative analysis (AQUA) of tissue microarrays. This method has been validated, has proved to be more accurate than pathologist-based scoring of brown stain (26, 27), and has been used in a number of prior melanoma studies (28–31). We found that expression of TRAIL receptors is increased in both primary and metastatic tumors compared with benign nevi, suggesting that TRAIL is a relevant therapeutic target in melanoma. High expression, defined arbitrarily as exceeding the 95th percentile of AQUA scores in benign nevi, was found in a substantially larger proportion of melanoma samples for TRAIL-R2 compared with TRAIL-R1. Our results indicate that assessment of baseline tumor receptor expression levels may be important in the analysis and interpretation of clinical trials involving TRAIL agonists and could possibly be used to select patients for treatment in future clinical trials.
Materials and Methods
Tissue microarray construction
The melanoma tissue microarrays were constructed as previously described (30). A total of 232 primary melanomas, 15 local recurrences, and 299 metastatic cores, each measuring 0.6 mm in diameter, were spaced 0.8 mm apart on glass slides. The cohort was constructed from paraffin-embedded, formalin-fixed tissue blocks obtained from the Yale University Department of Pathology Archives. Specimens and clinical information were collected under the guidelines and approval of a Yale University Institutional Review Board. The cohort has been used in prior publications (32). The specimens were resected between 1959 and 2000, with a follow-up range between 2 months and 40 years (mean follow-up time, 6.7 years). Age at diagnosis ranged from 18 to 91 years (mean age, 52.4 years). The cohort included 55% males and 45% females. The time between tumor resection and tissue fixation was not available. A pathologist reviewed slides from all of the blocks to select representative areas of invasive tumor to be cored. The cores were placed on the tissue microarray using a Tissue Microarrayer (Beecher Instruments, Silver Spring, MD). The tissue microarrays were then cut to 0.5-μm sections and placed on glass slides using an adhesive tape-transfer system (Instrumedics, Inc., Hackensack, NJ) with UV cross-linking. Similarly, a tissue microarray was made containing cores from 540 benign nevi. The nevi array contained 31 metastatic specimens from patients that were also represented on the melanoma array. Both arrays contained identical cell lines, cored from pellets as previously described (33). The overlapping metastatic specimens and cell lines were used for normalization of the scores obtained from the benign and malignant arrays.
Immunohistochemistry
Staining was done for automated analysis of melanoma specimens as previously described (29, 30). Briefly, slides were deparaffinized in xylene and transferred through two changes of 100% ethanol. For antigen retrieval, the slides were boiled in a pressure cooker containing 6.5 mmol/L sodium citrate (pH 6.0). Endogenous peroxidase activity was blocked in a mixture of methanol and 2.5% hydrogen peroxide for 30 minutes at room temperature. To reduce nonspecific background staining, slides were incubated at room temperature for 30 minutes in 0.3% bovine serum albumin/1× TBS. Slides were incubated at 4°C overnight in a humidity tray with the primary antibodies [rabbit polyclonal anti-DR5 immunoglobulin G at 1:350 (Oncogene Research, San Diego, CA) and mouse monoclonal anti-DR4 immunoglobulin G at 1:80 (R&D Systems, Inc., Minneapolis, MN)]. To create a tumor mask, slides were simultaneously incubated overnight with a primary anti-S100 antibody (rabbit anti-human S100 at 1:500 for DR4 and mouse anti-human S100 at 1:200 for DR5). Slides were rinsed thrice in 1× TBS/0.05% Tween 20. For TRAIL-R2, slides were incubated for 1 hour at room temperature with goat anti-rabbit horseradish peroxidase (Envision, DAKO Corp., Carpinteria, CA) to identify the target, and goat anti-mouse immunoglobulin G conjugated to Alexa 546 (Molecular Probes, Inc., Eugene, OR) at a dilution of 1:100 to identify the mask. The same technique was used to assess TRAIL-R1 expression, except that goat anti-mouse horseradish peroxidase and goat anti-rabbit immunoglobulin G conjugated to Alexa 546 were used. The slides were washed again as above and incubated for 10 minutes with Cy5 directly conjugated to tyramide (Perkin-Elmer, Boston, MA) at a dilution of 1:50 for primary antibody identification. The slides were rinsed again and coverslips were mounted with ProLong Gold antifade reagent, which contained 4,6-diamidine-2-phenylindole to identify the nuclei.
Automated image acquisition
Images were acquired using automated quantitative analysis as previously described (26, 27). Briefly, areas of tumor were distinguished from stroma by creating a mask with the S100 signal tagged with Alexa 546. Coalescence of S100 at the cell surface was used to identify the membrane/cytoplasm compartment within the tumor mask whereas 4,6-diamidino-2-phenylindole was used to identify the nuclear compartment within the tumor mask. The target markers, TRAIL-R1 and TRAIL-R2, were visualized with Cy5 (red). Cy5 was used because its emission peak is outside the color spectrum of tissue autofluorescence. Multiple monochromatic, high-resolution (1,024 × 1,024 pixel, 0.5-μm) grayscale images were obtained for each histospot using the 10× objective of an Olympus AX-51 epifluorescence microscope (Olympus, Melville, NY) with an automated microscope stage and digital image acquisition driven by custom program and macrobased interfaces with IPLabs software (Scanalytics, Inc., Fairfax, VA).
Algorithmic image analysis
Images were analyzed using algorithms that have previously been extensively described (26). Two images (one in-focus and one out-of-focus) were taken of the compartment specific tags and the target marker. A percentage of the out-of-focus image was subtracted from the in-focus image for each pixel, representing the signal-to-noise ratio of the image. An algorithm described as Rapid Exponential Subtraction Algorithm was used to subtract the out-of-focus information in a uniform fashion for the entire microarray. Subsequently, the Pixel Locale Assignment for Compartmentalization of Expression algorithm was used to assign each pixel in the image to a specific subcellular compartment and the signal in each location is calculated. Pixels that cannot accurately be assigned to a compartment were discarded. The data were saved and subsequently expressed as the average signal intensity per unit of compartment area. For the nuclear and membrane/cytoplasmic compartments, the image was measured on a scale of 0 to 255 and expressed as target signal intensity relative to the compartment area.
Statistical analysis
The Statview and JMP5 (SAS Institute, Inc., Cary, NC) software packages were used for data analyses. Continuous AQUA scores of target expression were divided by the median score and associations with clinical and pathologic variables were completed using the χ2 test. The prognostic significance of the variables was assessed for predictive value using the Cox proportional hazards model with overall survival as an end point. Survival curves were generated using the Kaplan-Meier method, with significance evaluated using the Mantel-Cox log-rank test. Comparison of expression in malignant and benign specimens, as well as comparisons between primary and metastatic specimens, was done with unpaired t tests.
Cell culture
Normal human melanocytes from newborn foreskins were grown as previously described (34). The cells were cultured in Ham’s F-12 medium (Life Technologies, Invitrogen Corp., Grand Island, NY) supplemented with 7% fetal bovine serum (Gemini Bio-Products, Woodland, CA), 100 units/mL penicillin, 100 μg/mL streptomycin (Life Technologies), termed basal medium, and enriched with 85 nmol/L 12-O-tetradecanoyl phorbol-13-acetate, 0.1 mmol/L 3-isobutyl-1-methyl-xanthine (IBMX), 2.5 nmol/L cholera toxin, 1 μmol/L Na3VO4, and 0.1 mmol/L N6,2′-O-dibutyry-ladenosine 3:5-cyclic monophosphate (all from Sigma-Aldrich, St. Louis, MO), termed TICVA, required for proliferation. Early-passage melanocyte cultures (passage 1–2) pooled from three to six Caucasian donors were used to reduce individual donor variations. The normal melanocytes proliferated at a rate of 3 to 4 day population doubling time and were highly differentiated and expressed all known melanocyte-specific genes.
The human primary melanoma cells WW165 (2.25-mm-deep lesion) were grown in OPTI-MEM (Life Technologies, Invitrogen) supplemented with 10% fetal bovine serum and antibiotics and enriched with IBMX required for optimal proliferation. The metastatic melanomas YUMAC (locally recurrent metastasis) and YUGEN8 (brain metastasis) were maintained in basal medium as described (35). YUDEW (subcutaneous metastasis), YULYO (cutaneous metastasis), YUFIC (lymph node metastasis), YUROB (distant soft-tissue metastasis), YUCAL (soft-tissue metastasis), and 1335 mel (lymph node metastasis) were grown in OPTI-MEM supplemented with 10% fetal bovine serum plus antibiotics. The normal melanocytes and the melanoma cell strains YUMAC, YUDEW, YULYO, YUFIC, YUROB, and YUCAL were harvested during the first or second passage in culture. The 501 Mel cells (obtained from Dr. Steven Rosenberg, Surgery Branch, National Cancer Institute, Bethesda, MD) were also maintained in basal medium.
The normal and malignant melanocytes were harvested during the logarithmic growth phase to avoid differences due to cell cycle distribution.
Western blot analysis
The cell pellets were lysed in radioimmuno-precipitation assay buffer (PBS, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS) and supplemented with a cocktail of protease inhibitors (“Complete” Boehringer Mannheim Corp., Roche Molecular Biochemicals, Indianapolis, IN). Total cell extracts (36 μg protein/lane), measured by the Bio-Rad protein assay (Bio-Rad laboratories, Hercules, CA), were fractionated in precast gels (10% polyacrylamide NuPAGE Bis-Tris, NOVEX, San Diego, CA) and Western blotted according to standard protocols. TRAIL-R1 was probed under nonreducing conditions (monoclonal antibody diluted 1:250) and TRAIL-R2 was probed under reducing conditions (antibodies diluted 1:500). Actin was probed (monoclonal antibody C-2, Santa Cruz Biotechnology, Santa Cruz, CA) and used as a measure for protein loading.
Results
To assess for intratumor heterogeneity, two separate sets of slides, each containing a core from a different area of the tumor for each patient, were used to evaluate the expression of each marker. Both receptors did not have significant amounts of nuclear staining in our specimens and only the membranous/cytoplasmic compartments were analyzed. We did not see differences in staining patterns within the tumor mask within a histospot. Using the Pearson correlation test, we found that the scores from matching spots on the two arrays were highly correlated (P < 0.0001) for both TRAIL-R1 and TRAIL-R2 expression. Figure 1 shows regression plots for the two arrays for TRAIL-R1 and TRAIL-R2 (R = 0.7 and R = 0.6, respectively). The vast majority of patients did not have high expression in one spot and low expression in another spot, as shown in this figure.
Fig. 1.
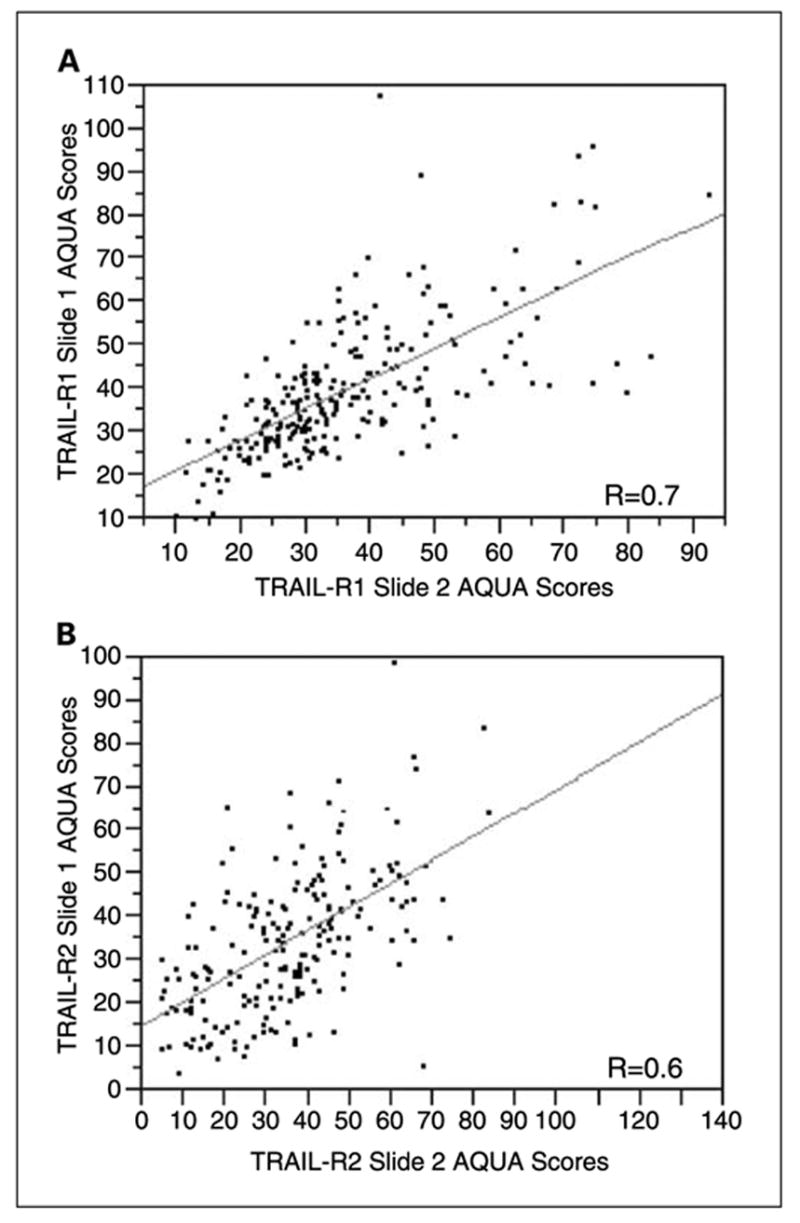
Regression plots for scores from the two sets of melanoma arrays for TRAIL-R1 (A) and TRAIL-R2 (B). Each array contains histospots from the same patients, taken from different areas of the tumor. Staining for TRAIL-R2 was slightly less homogeneous than for TRAIL-R1.
AQUA scores ranged from 5.919 to 88.545 for TRAIL-R1, with a median score of 34.769, and from 4.35 to 133.63 for TRAIL-R2, with a median score of 33.67. Examples of strong and weak TRAIL-R1 and TRAIL-R2 staining are shown in Fig. 2. Figure 2A to F shows staining with the different fluorophores: the top right quadrant of the histospots in these images show the nuclear staining using 4,6-diamidine-2-phenylindole; the top left quadrant shows S100 conjugated to Cy3 to reveal the tumor cells; the bottom left quadrant shows strong (A and E) or weak (B and F) staining for TRAIL-R1 and TRAIL-R2, respectively. The bottom right quadrant of these images is an artificially created merged image. The other images are high-magnification (×60) images of the TRAIL receptors. As can be seen in this figure, the staining was membranous and no significant nuclear staining was seen. A wide range of TRAIL receptor AQUA scores was seen. The strong TRAIL-R1 staining (C) corresponds to an AQUA score of 87.01, the weak TRAIL-R1 staining (D) to an AQUA score of 19.17. The strong TRAIL-R2 (G) staining shown corresponds to an AQUA score of 78.29, and the weak TRAIL-R2 (H) to an AQUA score of 13.64.
Fig. 2.
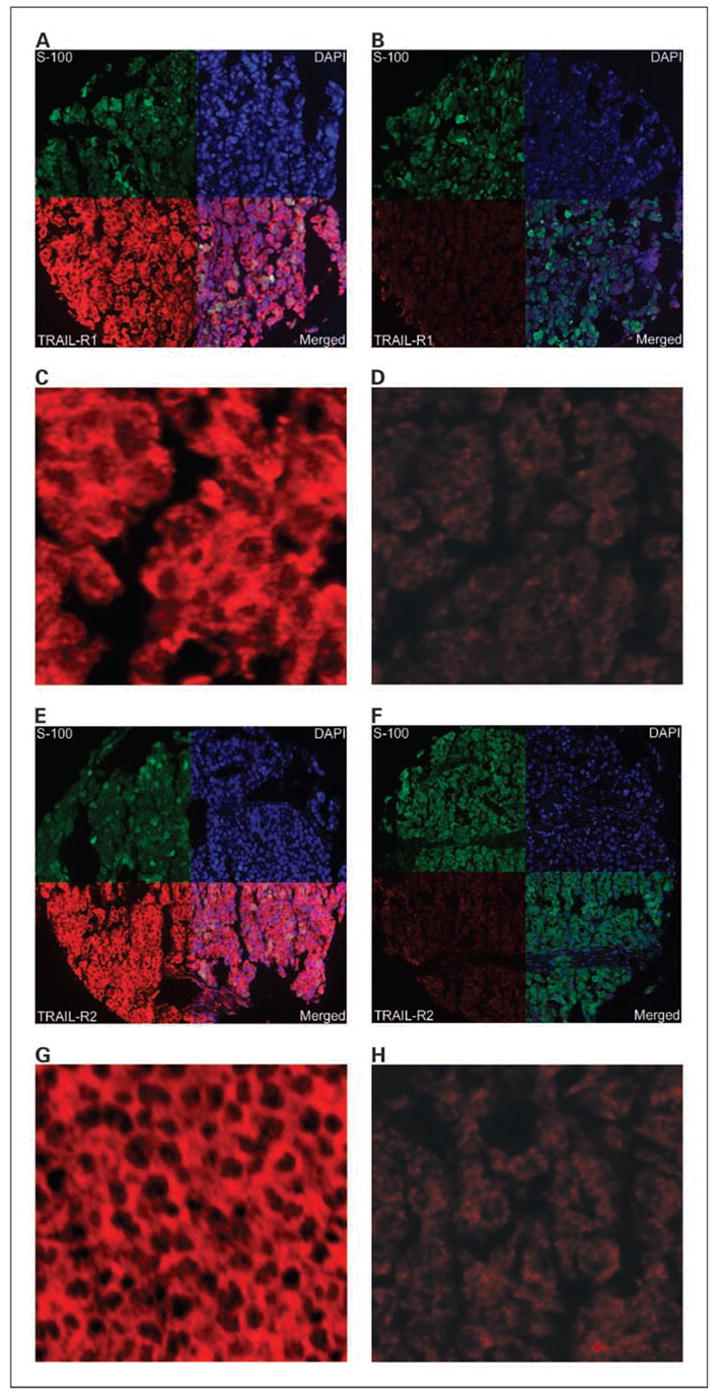
Membranous TRAIL-R1 and TRAIL-R2 expression in melanoma histospots using S100 to define the tumor mask (green), 4,6-diamidine-2-phenylindole to define the nuclear compartment (blue), and Cy5 (red) for the target (TRAIL-R1or TRAIL-R2). Merged images (A, B, E, and F, bottom right quadrants) show the amount of target within the nuclear compartment and within the cytoplasmic compartment within the tumor mask. High (A and C) and low (B and D) expression of TRAIL-R1at ×10 and ×60 magnifications, respectively, and high (E and G) and low (F and H) expression of TRAIL-R2 at ×10 and ×60 magnifications, respectively. TRAIL-R1expression in (C and D) corresponds to AQUA scores of 87.01 and 19.171, respectively. TRAIL-R2 expression in (G and H) corresponds to AQUA scores of 78.29 and 13.64, respectively.
For each of the two markers, the AQUA scores from both sets of slides were combined to give a single data set. Of the 546 melanoma histospots on each slide, 228 were interpretable for both cores for TRAIL-R1 and 245 were interpretable for one core. Tumor spots were deemed uninterpretable if they had insufficient tumor cells, loss of tissue in the spot, or an abundance of necrotic tissue. For patients who had two interpretable histospots, a composite score was formed by taking the average of the two scores. For patients with only one interpretable core, the single score was used. The combined data set for TRAIL-R1 had scores for 473 melanoma patients. For TRAIL-R2, 203 patients had two scores and 238 had one score, yielding a data set with scores for 441 melanoma patients. These numbers are summarized in Table 1. For the nevus specimens, we obtained 362 scores for TRAIL-R1 and 398 scores for TRAIL-R2.
Table 1.
Histospots interpretable for TRAIL-R1 and TRAIL-R2 staining
| Melanomas with two scores | Melanomas with one score | Total melanomas with AQUA scores | |
|---|---|---|---|
| TRAIL-R1 | 228 | 245 | 473 |
| TRAIL-R2 | 203 | 238 | 441 |
Because TRAIL-R1 and TRAIL-R2 are attractive drug targets, we assessed the differences between benign and malignant tissues. Unpaired t tests showed that expression was significantly higher in malignant versus benign tissue cores for both receptors (P < 0.0001 for both), as shown in Fig. 3. However, the differences in expression were noticeably larger for TRAIL-R2.
Fig. 3.
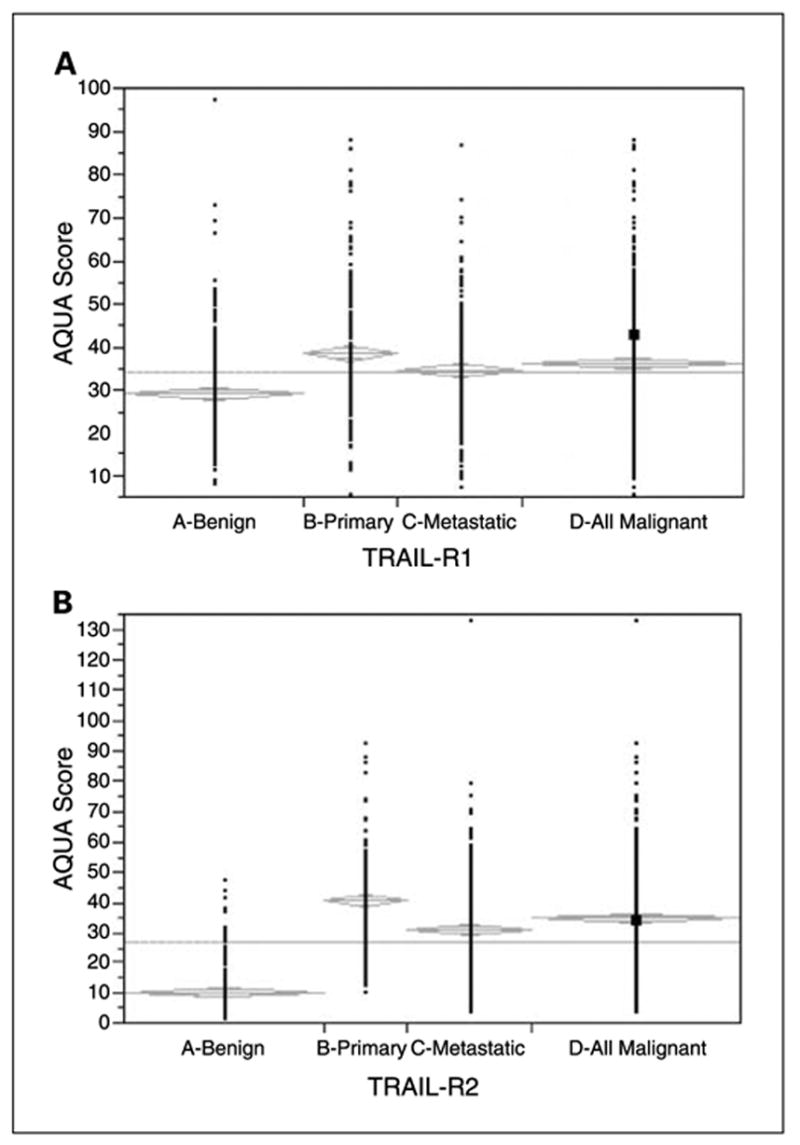
Comparison of TRAIL-R1 (A) and TRAIL-R2 (B) expression in nevi, primary melanomas, metastatic melanomas, and all malignant specimens.
Using the Cox univariate survival analysis of raw AQUA scores, we found that TRAIL-R1 expression was not associated with melanoma-specific survival (P = 0.5014). Although high TRAIL-R2 expression was correlated with increased survival in the entire cohort (P = 0.0439), when breaking down the patient cohorts into primary and metastatic specimens, there was no association with survival for either TRAIL-R1 or TRAIL-R2 in primary or metastatic subsets. Using the Cox proportional hazards model, we did multivariate analysis to assess the independent predictive value of TRAIL-R2 expression. TRAIL-R2 expression did not retain its independent predictive value as it was associated with disease stage (primary versus metastatic).
To assess the association between TRAIL receptor expression and other commonly used clinical and pathologic variables, we used the χ2 test, dividing scores by the median expression for malignant specimens. Both TRAIL-R1 and TRAIL-R2 expressions were strongly associated with disease stage; expression of both receptors was significantly higher in primary specimens than in metastatic specimens by unpaired t tests (P = 0.0031 for TRAIL-R1; P < 0.0001 for TRAIL-R2). We found no association between TRAIL-R1 or TRAIL-R2 and age, gender, Breslow depth, Clark’s level, and presence of ulceration.
Preclinical data indicated that tumor cell sensitivity to TRAIL-induced apoptosis was associated with receptor expression level. Although our assay does not provide information on the threshold expression level for sensitivity to TRAIL agonists, we nevertheless felt that we could classify tumors as high or low expressers, based on the range of expression in our very large cohort of benign nevi, and the assumption that levels of expression in benign nevi are low from a biological perspective. AQUA gives continuous output data, as opposed to brown stain, which is read as “positive” or “negative.” Here we define “low” or “normal” AQUA scores as those that fall at or below the 95th percentile score for nevi for each marker. This score was 47 for TRAIL-R1 and 22.7 for TRAIL-R2, and scores above these cutoffs in the malignant specimens were considered “high.” As shown in Fig. 4, a larger fraction of malignant specimens had high TRAIL-R2 expression than TRAIL-R1 (74% and 19% respectively). When using this definition of high expression, 26% of primary specimens and 14% of metastatic specimens had high TRAIL-R1 expression whereas 84% of primary and 67% of metastatic specimens had high TRAIL-R2 expression, suggesting that TRAIL-R2 might be a better drug target in melanoma.
Fig. 4.
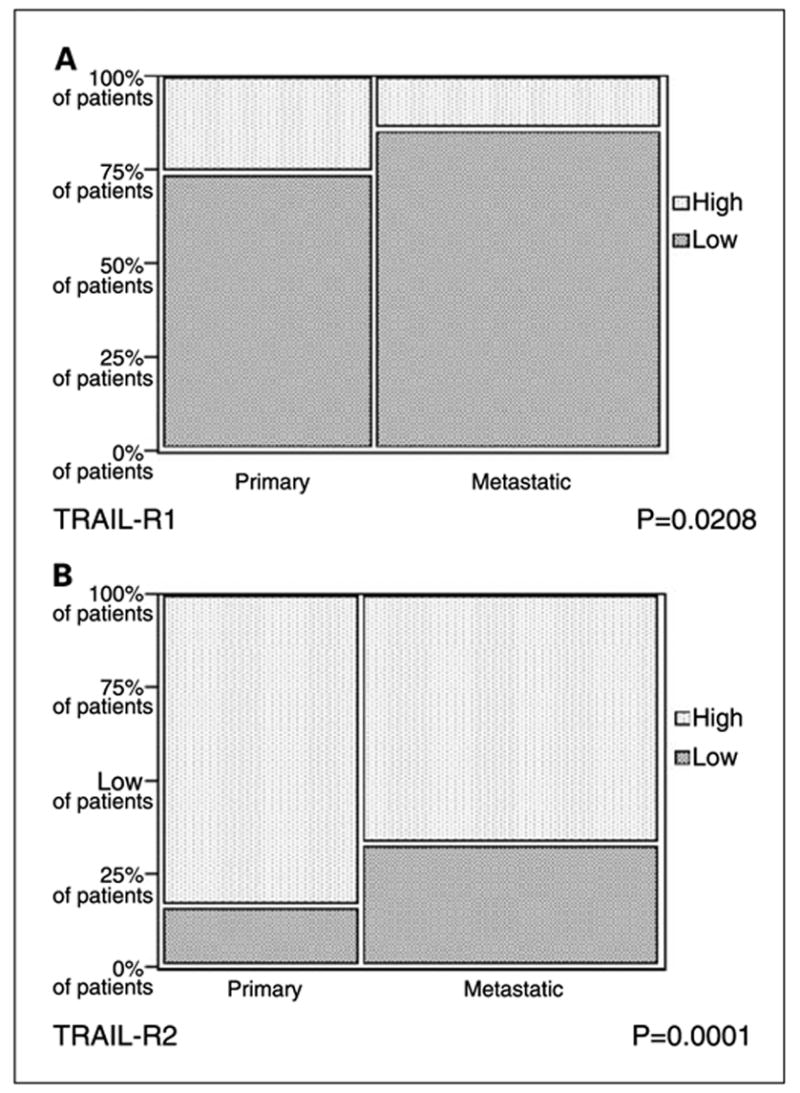
χ2 analysis comparing high and normal/low TRAIL-R1 (A) and TRAIL-R2 (B) in primary and metastatic melanoma specimens. We defined high TRAIL receptor expression for malignant specimens as expression that was higher than the 95th percentile score for nevi. The cutoff score was 47 for TRAIL-R1and 22.7 for TRAIL-R2.
To assess the degree of coexpression of TRAIL-R1 and TRAIL-R2, we did a regression analysis and found a weak association between the two receptors (R = 0.25). Using the above definition of high expression based on the 95th percentile score for nevi, 23% of patients with high TRAIL-R2 expression also had high TRAIL-R1 expression and 93% of patients with high TRAIL-R1 expression also had high TRAIL-R2 expression. Twenty-six percent of patients did not have high expression of either receptor.
Western blots using a limited number of cell strains confirmed general up-regulation of both receptors in melanoma cells compared with normal melanocytes. Probing for TRAIL-R1 revealed a broad band due to the glycosylated forms of the receptor. The level of this receptor was low in normal melanocytes and, with the exception of YUGEN8, high levels were seen in the malignant cells. TRAIL-R2 was lower in the normal melanocytes than in most of the malignant cells (Fig. 5). To account for uneven loading, expression had been normalized to the actin level in normal melanocytes.
Fig. 5.
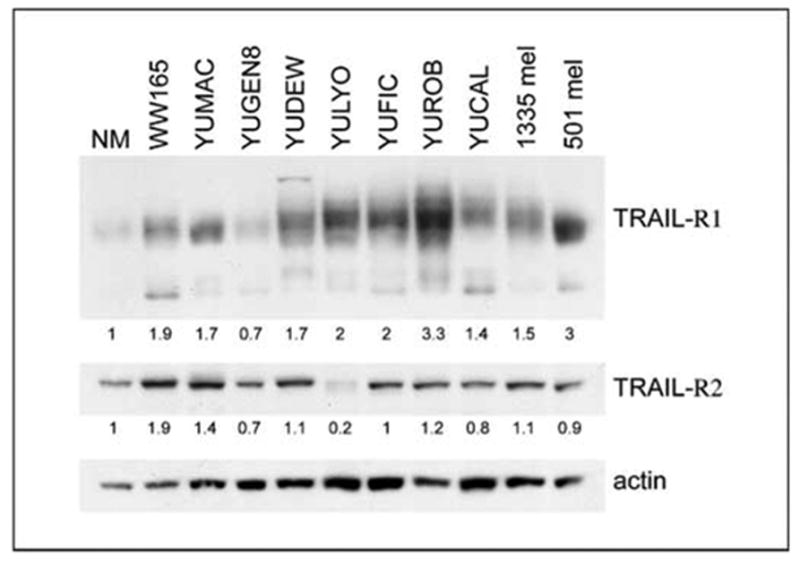
TRAIL receptor expressions in normal and malignant melanocytes. Western blot of extracts derived from normal melanocytes (NM) and several melanoma cell strains (as indicated) employing anti – TRAIL-R1and anti – TRAIL-R2 antibodies. TRAIL-R1appeared as a broad band of about 40 to 60 kDa that could represent different glycosylated forms of the receptor. The lower molecular weight interactive bands may represent unglycosylated forms or degradation products of the receptor. TRAIL-R2 appeared as a protein species of ~55 to 60 kDa. Actin was used as a loading control. The numbers show the ratios of band intensity in the melanoma cell strains relative to that in normal melanocytes, normalized to the actin loading.
Discussion
The purpose of this study was to assess the expression of TRAIL-R1 and TRAIL-R2 in a quantitative fashion on a large cohort of melanoma specimens in an objective, automated fashion, and to compare expression of these receptors in malignant specimens to expression in benign nevi. Furthermore, we evaluated the association between TRAIL receptor expression and clinical and pathologic variables. Expression of both TRAIL-R1 and TRAIL-R2 was significantly higher in malignant than in benign specimens and significantly higher in primary melanomas than in metastatic specimens. TRAIL-R1 and TRAIL-R2 expression was not associated with improved survival in the primary or metastatic cohorts. Our results were reproducible when using a second array with different cores from the same patients, with little intratumor variability. Our immunohistochemical results corresponded to the higher expression by Western blots using a limited number of samples in most, but not all, of the patient-derived cell strains compared with normal melanocytes.
Ligand binding to TRAIL-R1 and TRAIL-R2 induces apoptosis; hence, finding higher expression in melanoma than in nevi was encouraging for development of TRAIL-targeted therapeutics in melanoma. However, there was substantial overlap in the range of expression between benign nevi and melanoma, particularly for TRAIL-R1. Because level of expression may be an important determinant of sensitivity to TRAIL agonists, we sought to determine the percent of samples that displayed receptor overexpression as compared with benign nevi. For this purpose, we arbitrarily defined high expression as exceeding the AQUA score in 95% of benign nevi. We found substantial differences for TRAIL-R1 versus TRAIL-R2; for TRAIL-R1, only 26% and 14% of primary and metastatic melanoma samples, respectively, met our definition for high expression, whereas for TRAIL-R2, 84% and 67%, respectively, were considered high expressers. We found a weak association between TRAIL-R1 and TRAIL-R2 expression, and 26% of patients did not have high expression of either receptor. These results may have important implications for design of clinical trials using these agents and suggest that pretreatment assessment for receptor expression should be incorporated into the studies, at the very least for analysis of outcome. The results also suggest that targeting TRAIL-R2 may be more effective in melanoma.
Markers that have large differences in expression between benign and malignant melanocytes can be of clinical diagnostic utility. Lesions such as atypical nevi or Spitz nevi are often difficult to differentiate from melanomas. Whereas differences in expression of TRAIL-R2 between nevi and melanoma specimens were significant, there is enough overlap to preclude use of TRAIL-R2 expression as a single diagnostic marker. However, it might be valuable as a future diagnostic marker if used in combination with other discriminating markers.
Although high TRAIL receptor expression was associated with disease stage, we found no correlation between level of expression and survival in the primary or metastatic cohorts. In similar studies done in breast cancer by our group (36), there was an inverse correlation between TRAIL-R2 expression and survival, possibly because high TRAIL-R expression may reflect activity of the antiapoptotic nuclear factor κB pathway. The results in both the melanoma and breast cancer cohorts are consistent with the complex interactions between the TRAIL receptors, external factors such as immune cell infiltrates, and with the intracellular apoptotic machinery. For example, coexpression by tumor cells of the TRAIL ligand may influence outcome related to expression of the TRAIL receptors. Expression of TRAIL ligand has been assessed in a small study of 45 primary cutaneous melanomas, and high TRAIL expression on melanoma cells was found to be associated with a high mitotic rate but not with other staging variables (37). Coexpression of decoy receptors may also influence outcome, although in prior in vitro studies, presence or absence of decoy receptors did not seem to affect sensitivity to TRAIL-R antibodies or to TRAIL and signals transduced through either of the two death receptors were sufficient to induce apoptosis (38). Intracellular mechanisms of resistance to TRAIL-induced apoptosis are also likely to influence outcome in patients whose tumors express TRAIL-receptors. The mechanism of resistance to TRAIL-induced apoptosis in melanoma is complex. High levels of FLICE-inhibitory protein have been shown in TRAIL-resistant melanomas and low levels for TRAIL-sensitive melanomas (23, 39). Agents that decrease FLICE-inhibitory protein expression have been shown to sensitize melanoma and other cells to TRAIL (38, 39). Additional antiapoptotic members of the apoptotic pathways have been implicated in resistance to TRAIL-induced apoptosis, such as X-linked inhibitor of apoptosis and survivin (40, 41). Other researchers have shown that melanoma cell lines that have lost their TRAIL sensitivity proliferate faster than their parental cell lines, and this is associated with reduced p53 and p21 expression and increased activation of Erk and Akt (42). Resistance of melanoma cells to TRAIL-induced apoptosis is also associated with nuclear factor κB activation (43), and inhibition of nuclear factor κB and antiapoptotic events downstream of nuclear factor κB can reverse this resistance (21, 44).
In summary, our studies show stronger TRAIL-R1 and TRAIL-R2 expression in melanocytic tumors than in benign nevus specimens, although substantial overlap in the range of expression was observed, particularly for TRAIL-R1. For TRAIL-R2, the expression in the majority of melanoma samples exceeded the 95th percentile for benign nevi. These data indicate that assessing receptor levels and tumor heterogeneity should be included in future clinical trials using TRAIL receptor–activating agents for treatment of melanoma. Because preclinical work on melanoma cells indicate that TRAIL receptor expression is associated with induction of a response by agents that activate these receptors (23–25), our work also suggests that TRAIL-R2 might be a superior therapeutic target in melanoma to TRAIL-R1. Further work is needed to elucidate the biological significance of TRAIL-R1 and TRAIL-R2 expression and to establish the association between TRAIL receptor expression and response to TRAIL receptor agonists.
Footnotes
Note: M.M. McCarthy and K.A. DiVito contributed equally to this work.
Grant support: Progetto Italia-U.S.A. grant 530/F-A1 (D. Kovacs); NIH grant K0-8 ES11571and the Breast Cancer Alliance (R.L. Camp); the Patrick and Catherine Weldon Donaghue Medical Research Foundation, NIH Department of Defense grant NIHR21CA100825-01 (D.L. Rimm); and NIH grants R0-1CA115756-01and P30 041942 and the Breast Cancer Alliance (H.M. Kluger).
References
- 1.Rigel DS. Malignant melanoma: incidence issues and their effect on diagnosis and treatment in the 1990s. Mayo Clin Proc. 1997;72:367–71. doi: 10.4065/72.4.367. [DOI] [PubMed] [Google Scholar]
- 2.Chapman PB, Einhorn LH, Meyers ML, et al. Phase III multicenter randomized trial of the Dartmouth regimen versus dacarbazine in patients with metastatic melanoma. J Clin Oncol. 1999;17:2745–51. doi: 10.1200/JCO.1999.17.9.2745. [DOI] [PubMed] [Google Scholar]
- 3.Falkson CI, Ibrahim J, Kirkwood JM, Coates AS, Atkins MB, Blum RH. Phase III trial of dacarbazine versus dacarbazine with interferon α-2b versus dacarbazine with tamoxifen versus dacarbazine with interferon α-2b and tamoxifen in patients with metastatic malignant melanoma: an Eastern Cooperative Oncology Group study. J Clin Oncol. 1998;16:1743–51. doi: 10.1200/JCO.1998.16.5.1743. [DOI] [PubMed] [Google Scholar]
- 4.Tsao H, Atkins MB, Sober AJ. Management of cutaneous melanoma. N Engl J Med. 2004;351:998–1012. doi: 10.1056/NEJMra041245. [DOI] [PubMed] [Google Scholar]
- 5.Motoki K, Mori E, Matsumoto A, et al. Enhanced apoptosis and tumor regression induced by a direct agonist antibody to tumor necrosis factor-related apoptosis-inducing ligand receptor 2. Clin Cancer Res. 2005;11:3126–35. doi: 10.1158/1078-0432.CCR-04-1867. [DOI] [PubMed] [Google Scholar]
- 6.Pukac L, Kanakaraj P, Humphreys R, et al. HGS-ETR1, a fully human TRAIL-receptor 1monoclonal antibody, induces cell death in multiple tumour types in vitro and in vivo. Br J Cancer. 2005;92:1430–41. doi: 10.1038/sj.bjc.6602487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelley SK, Ashkenazi A. Targeting death receptors in cancer with Apo2L/TRAIL. Curr Opin Pharmacol. 2004;4:333–9. doi: 10.1016/j.coph.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science. 1998;281:1305–8. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 9.Kelley RF, Totpal K, Lindstrom SH, et al. Receptor-selective mutants of apoptosis-inducing ligand 2/tumor necrosis factor-related apoptosis-inducing ligand reveal a greater contribution of death receptor (DR) 5 than DR4 to apoptosis signaling. J Biol Chem. 2005;280:2205–12. doi: 10.1074/jbc.M410660200. [DOI] [PubMed] [Google Scholar]
- 10.Kurbanov BM, Geilen CC, Fecker LF, Orfanos CE, Eberle J. Efficient TRAIL-R1/DR4-mediated apoptosis in melanoma cells by tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) J Invest Dermatol. 2005;125:1010–9. doi: 10.1111/j.0022-202X.2005.23900.x. [DOI] [PubMed] [Google Scholar]
- 11.Ashkenazi A, Pai RC, Fong S, et al. Safety and anti-tumor activity of recombinant soluble Apo2 ligand. J Clin Invest. 1999;104:155–62. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keane MM, Ettenberg SA, Nau MM, Russell EK, Lipkowitz S. Chemotherapy augments TRAIL-induced apoptosis in breast cell lines. Cancer Res. 1999;59:734–41. [PubMed] [Google Scholar]
- 13.Singh TR, Shankar S, Chen X, Asim M, Srivastava RK. Synergistic interactions of chemotherapeutic drugs and tumor necrosis factor-related apoptosis-inducing ligand/Apo-2 ligand on apoptosis and on regression of breast carcinoma in vivo. Cancer Res. 2003;63:5390–400. [PubMed] [Google Scholar]
- 14.Ballestrero A, Nencioni A, Boy D, et al. Tumor necrosis factor-related apoptosis-inducing ligand cooperates with anticancer drugs to overcome chemoresistance in antiapoptotic Bcl-2 family members expressing Jurkat cells. Clin Cancer Res. 2004;10:1463–70. doi: 10.1158/1078-0432.ccr-1365-02. [DOI] [PubMed] [Google Scholar]
- 15.Mitsiades CS, Treon SP, Mitsiades N, et al. TRAIL/ Apo2L ligand selectively induces apoptosis and overcomes drug resistance in multiple myeloma: therapeutic applications. Blood. 2001;98:795–804. doi: 10.1182/blood.v98.3.795. [DOI] [PubMed] [Google Scholar]
- 16.Shankar S, Chen X, Srivastava RK. Effects of sequential treatments with chemotherapeutic drugs followed by TRAIL on prostate cancer in vitro and in vivo. Prostate. 2005;62:165–86. doi: 10.1002/pros.20126. [DOI] [PubMed] [Google Scholar]
- 17.Nakata S, Yoshida T, Horinaka M, Shiraishi T, Wakada M, Sakai T. Histone deacetylase inhibitors up-regulate death receptor 5/TRAIL-R2 and sensitize apoptosis induced by TRAIL/APO2-L in human malignant tumor cells. Oncogene. 2004;23:6261–71. doi: 10.1038/sj.onc.1207830. [DOI] [PubMed] [Google Scholar]
- 18.Nyormoi O, Mills L, Bar-Eli M. An MMP-2/MMP-9 inhibitor, 5a, enhances apoptosis induced by ligands of the TNF receptor superfamily in cancer cells. Cell Death Differ. 2003;10:558–69. doi: 10.1038/sj.cdd.4401209. [DOI] [PubMed] [Google Scholar]
- 19.Chuntharapai A, Dodge K, Grimmer K, et al. Isotype-dependent inhibition of tumor growth in vivo by monoclonal antibodies to death receptor 4. J Immunol. 2001;166:4891–8. doi: 10.4049/jimmunol.166.8.4891. [DOI] [PubMed] [Google Scholar]
- 20.Buchsbaum DJ, Zhou T, Grizzle WE, et al. Antitumor efficacy of TRA-8 anti-DR5 monoclonal antibody alone or in combination with chemotherapy and/or radiation therapy in a human breast cancer model. Clin Cancer Res. 2003;9:3731–41. [PubMed] [Google Scholar]
- 21.Nguyen T, Thomas W, Zhang XD, Gray C, Hersey P. Immunologically-mediated tumour cell apoptosis: the role of TRAIL in T cell and cytokine-mediated responses to melanoma. Forum (Genova) 2000;10:243–52. [PubMed] [Google Scholar]
- 22.Thomas WD, Hersey P. TNF-related apoptosis-inducing ligand (TRAIL) induces apoptosis in Fas ligand-resistant melanoma cells and mediates CD4 T cell killing of target cells. J Immunol. 1998;161:2195–200. [PubMed] [Google Scholar]
- 23.Griffith TS, Chin WA, Jackson GC, Lynch DH, Kubin MZ. Intracellular regulation of TRAIL-induced apoptosis in human melanoma cells. J Immunol. 1998;161:2833–40. [PubMed] [Google Scholar]
- 24.Zhang XD, Franco A, Myers K, Gray C, Nguyen T, Hersey P. Relation of TNF-related apoptosis-inducing ligand (TRAIL) receptor and FLICE-inhibitory protein expression to TRAIL-induced apoptosis of melanoma. Cancer Res. 1999;59:2747–53. [PubMed] [Google Scholar]
- 25.Nguyen T, Zhang XD, Hersey P. Relative resistance of fresh isolates of melanoma to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis. Clin Cancer Res. 2001;7:966–73s. [PubMed] [Google Scholar]
- 26.Camp RL, Chung GG, Rimm DL. Automated sub-cellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–7. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 27.Rubin MA, Zerkowski MP, Camp RL, et al. Quantitative determination of expression of the prostate cancer protein α-methylacyl-CoA racemase using automated quantitative analysis (AQUA): a novel paradigm for automated and continuous bio-marker measurements. Am J Pathol. 2004;164:831–40. doi: 10.1016/s0002-9440(10)63171-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–22. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 29.Divito KA, Berger AJ, Camp RL, Dolled-Filhart M, Rimm DL, Kluger HM. Automated quantitative analysis of tissue microarrays reveals an association between high Bcl-2 expression and improved outcome in melanoma. Cancer Res. 2004;64:8773–7. doi: 10.1158/0008-5472.CAN-04-1387. [DOI] [PubMed] [Google Scholar]
- 30.Berger AJ, Camp RL, Divito KA, Kluger HM, Halaban R, Rimm DL. Automated quantitative analysis of HDM2 expression in malignant melanoma shows association with early-stage disease and improved outcome. Cancer Res. 2004;64:8767–72. doi: 10.1158/0008-5472.CAN-04-1384. [DOI] [PubMed] [Google Scholar]
- 31.Berger AJ, Davis DW, Tellez C, et al. Automated quantitative analysis of activator protein-2α subcellular expression in melanoma tissue microarrays correlates with survival prediction. Cancer Res. 2005;65:11185–92. doi: 10.1158/0008-5472.CAN-05-2300. [DOI] [PubMed] [Google Scholar]
- 32.Kluger HM, DiVito K, Berger AJ, et al. Her2/neu is not a commonly expressed therapeutic target in melanoma—a large cohort tissue microarray study. Melanoma Res. 2004;14:207–10. doi: 10.1097/01.cmr.0000130874.33504.2f. [DOI] [PubMed] [Google Scholar]
- 33.Kluger HM, Kluger Y, Gilmore-Hebert M, et al. cDNA microarray analysis of invasive and tumorigenic phenotypes in a breast cancer model. Lab Invest. 2004;84:320–31. doi: 10.1038/labinvest.3700044. [DOI] [PubMed] [Google Scholar]
- 34.Hoek K, Rimm DL, Williams KR, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–82. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 35.von Willebrand M, Zacksenhaus E, Cheng E, Glazer P, Halaban R. The tyrphostin AG1024 accelerates the degradation of phosphorylated forms of retinoblastoma protein (pRb) and restores pRb tumor suppressive function in melanoma cells. Cancer Res. 2003;63:1420–9. [PubMed] [Google Scholar]
- 36.McCarthy MM, Sznol M, DiVito KA, Camp RL, Rimm DL, Kluger HM. Evaluating the expression and prognostic value of TRAIL-R1and TRAIL-R2 in breast cancer. Clin Cancer Res. 2005;11:5188–94. doi: 10.1158/1078-0432.CCR-05-0158. [DOI] [PubMed] [Google Scholar]
- 37.Bron LP, Scolyer RA, Thompson JF, Hersey P. Histological expression of tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) in human primary melanoma. Pathology. 2004;36:561–5. doi: 10.1080/00313020400011268. [DOI] [PubMed] [Google Scholar]
- 38.Griffith TS, Rauch CT, Smolak PJ, et al. Functional analysis of TRAIL receptors using monoclonal antibodies. J Immunol. 1999;162:2597–605. [PubMed] [Google Scholar]
- 39.Xiao C, Yang BF, Song JH, Schulman H, Li L, Hao C. Inhibition of CaMKII-mediated c-FLIP expression sensitizes malignant melanoma cells to TRAIL-induced apoptosis. Exp Cell Res. 2005;304:244–55. doi: 10.1016/j.yexcr.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 40.Zhang XD, Zhang XY, Gray CP, Nguyen T, Hersey P. Tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis of human melanoma is regulated by smac/DIABLO release from mitochondria. Cancer Res. 2001;61:7339–48. [PubMed] [Google Scholar]
- 41.Grossman D, Kim PJ, Schechner JS, Altieri DC. Inhibition of melanoma tumor growth in vivo by survivin targeting. Proc Natl Acad Sci U S A. 2001;98:635–40. doi: 10.1073/pnas.230450097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu JJ, Zhang XD, Gillespie S, Hersey P. Selection for TRAIL resistance results in melanoma cells with high proliferative potential. FEBS Lett. 2005;579:1940–4. doi: 10.1016/j.febslet.2005.02.041. [DOI] [PubMed] [Google Scholar]
- 43.Franco AV, Zhang XD, Van Berkel E, et al. The role of NF-κB in TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis of melanoma cells. J Immunol. 2001;166:5337–45. doi: 10.4049/jimmunol.166.9.5337. [DOI] [PubMed] [Google Scholar]
- 44.Chawla-Sarkar M, Bauer JA, Lupica JA, et al. Suppression of NF-κB survival signaling by nitrosyl-cobalamin sensitizes neoplasms to the anti-tumor effects of Apo2L/TRAIL. J Biol Chem. 2003;278:39461–9. doi: 10.1074/jbc.M306111200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]


