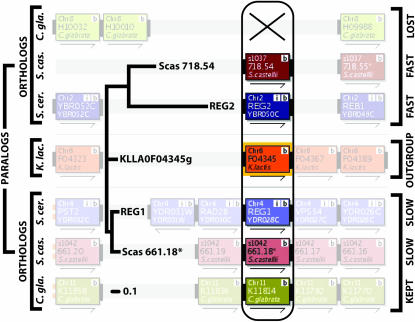
Figure 2.—
The REG1/REG2 locus. The YGOB (Byrne and Wolfe 2005) screenshot in the background shows the syntenic context at the locus, identifying orthologs and paralogs. The topology of the maximum-likelihood tree, from the Scer–Scas comparison in the 2:2 category, agrees perfectly with the topology derived from synteny, so this locus passes our phylogenetic filter and is retained in set 2. Furthermore, the orthologs REG2 and Scas_718.54 are the faster-evolving ohnologs in each species, making this a locus with consistent asymmetric evolution. The “X” marks the loss of the “fast” copy of the gene from C. glabrata, when it is compared to either S. cerevisiae (Scer–Cgla, 2:1) or S. castellii (Cgla–Scas, 1:2). Branch lengths show the pronounced asymmetric protein sequence evolution on both the shared and the terminal branches for this locus.