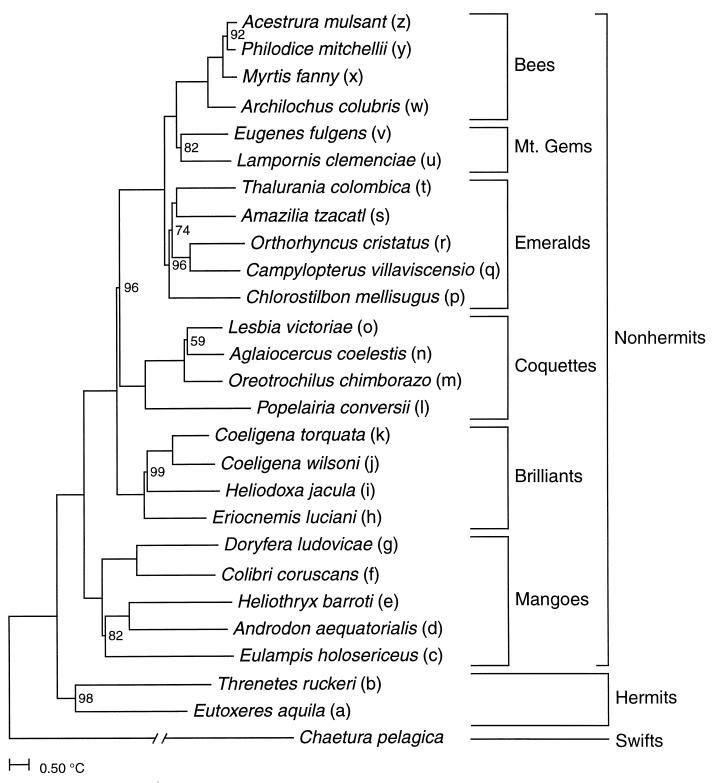
Figure 1.
Consensus unweighted least-squares FITCH topology obtained (8) from a complete matrix of symmetrized ΔTmH-C values rooted with the outgroup swift Chaetura pelagica; names refer to principal nonhermit lineages and relevant subfamilies and families of the Apodiformes (hummingbirds and swifts), and letter codes to species as plotted in Fig. 2. The ΔTmH-C index was obtained through several steps that minimize inaccuracies in distance measures (8). First, the T50H index was obtained by correcting raw median melting temperatures (Tm) for normalized percentage hybridization (NPH) through application of the second-order polynomial found to fit observed values of T50H regressed on Tm so as to avoid the excessive experimental error inherent in raw measures of NPH. The resulting T50H values were multiplied by the empirically determined scaling factor of 1.2 for percentage sequence divergence (27) and then corrected for homoplasy (28). Finally, these distances were converted to so-called delta (Δ) values by standardizing the melting temperatures of different-species (heterologous) hybrids to the melting temperatures of same-species (homologous) hybrids (8). After symmetrization (29), average path lengths (12) for the resulting ΔTmH-C values were estimated from 1,000 unweighted least-squares FITCH topologies (30) generated for a corresponding number of bootstrap pseudoreplicate matrices drawn from the complete matrix of 2,025 reciprocal genetic distances (three, rarely fewer, replicates per comparison). Internode support as indicated by bootstrap percentages (out of 1,000, if <100%) suggests strong support for the symmetrized topology, which was stable to multiple-deletion jackknifing (12). [Reproduced with permission from ref. 8. (Copyright 1997, Society for Molecular Biology and Evolution).]