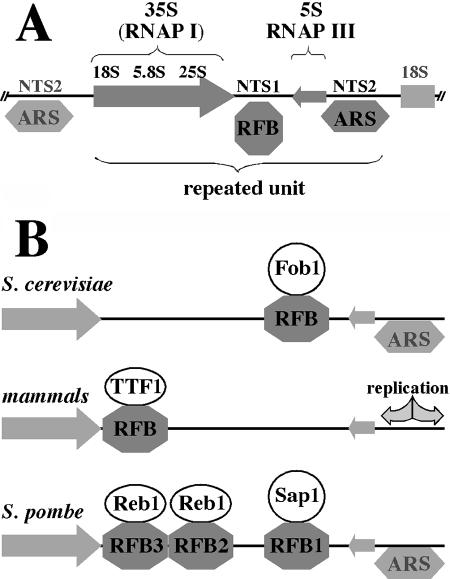
FIG. 2.
Ribosomal RFBs. (A) Organization of rRNA locus in S. cerevisiae. The locus consists of multiple units of direct repeats, each of which contains a 35S RNA transcription unit (which after processing gives rise to 18S, 5.8S, and 25S rRNAs) transcribed by RNA polymerase I, a 5S RNA gene transcribed by RNA polymerase III, and two nontranscribed spacers. One nontranscribed spacer (NTS2) contains a bidirectional origin of replication (ARS), whereas the other nontranscribed spacer (NTS1) contains an RFB that arrests forks that are about to enter the 35S transcription unit (i.e., forks that move from right to left). (B) Comparison of ribosomal RFBs in budding yeast, mammals, and fission yeast. In budding yeast, the RFB is caused by the Fob1 protein; in mammals, the RFB is caused by the TTF-1 protein, which is also a transcription termination factor for rRNAs. Fission yeast combines both scenarios: RFB1 is caused by Sap1, which, like Fob1, is not involved in termination of rRNA transcription, and RFB2 and RFB3 are caused by Reb1, which, like TTF-1, is a transcription termination factor. Arrows, rRNA transcription units. The fork that moves from right to left is blocked.