Figure 1.
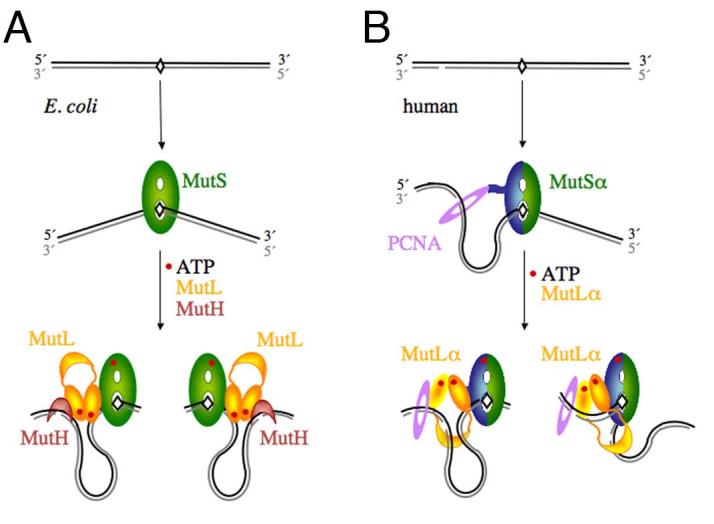
Diagrams of the initial incision step of MMR in E.coli and humans. A. In E. coli, MutS recognizes a mismatch, binds it and bends the DNA by 60° towards the major groove. The newly synthesized and unmethylated daughter strand is shown in grey. In the presence of ATP, MutL is recruited to the MutS-mismatch complex, and together they activate MutH to nick the daughter strand on either the 5′ or 3′ side of the mismatch. Since the DNA binding activity of MutL is not necessary for this step and slightly inhibits MutH activation [34], we propose that the DNA in between the mismatch and incision site is looped out. B. In humans, MutSα is made of MSH2 (green) and MSH6 (blue) and interacts with PCNA (purple). The daughter strand is marked by a pre-existing strand break. A break 3′ to the mismatch is shown here as an example. MutSα may have a strand preference when loaded by PCNA. The ensuing MutSα-MutLα-mismatch complex could also be biased due to the asymmetric (heterodimeic) nature. Incisions by the PMS2 subunit of MutLα on the 3′ side of the mismatch may be guided by PCNA and MutSα, and incisions on the 5′ side may be limited by how far the C-terminal domain of MutLα (the endonuclease active site) can extend from the MutSα-MutLα complex located on the mismatch site.
