Figure 2.
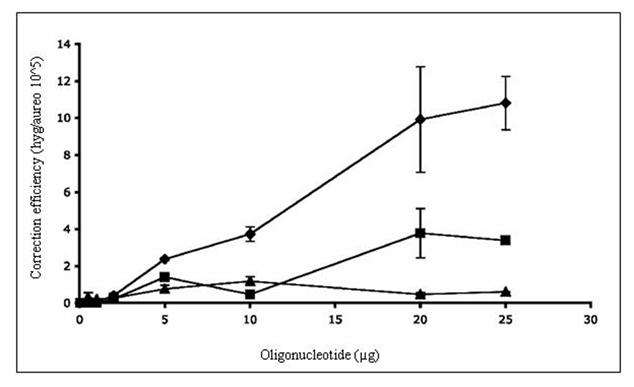
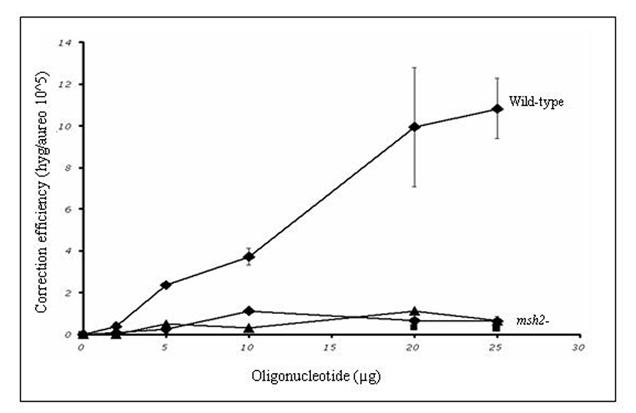
TNE of a chromosomal frameshift mutation in wild-type and mutant yeast. A) The yeast strain LSY678 (MATa leu2-3,112 trp1-1 ura3-1 his3-11,15 ade2-1 can1-100) was used as a host to demonstrate the repair of a chromosomal frameshift mutation. The oligonucleotides Hyg3S/74NT-A (◆), Hyg/3S74T-T (■), or Hyg/3S47NT-A (▲) were introduced over a range of concentrations into the strain by electroporation and after 16 hours of recovery the cells were plated on selective media to assess the level of correction. The correction efficiency (CE) is determined by the number of hygromycin resistant colonies (corrected cells) divided by the number of aureobasidin colonies (total number of viable cells capable of being corrected). B) The targeting oligonucleotides Hyg3S/74NT-A, Hyg3S/47NT-A, and Hyg3S/74T-T were delivered at a range of concentrations to LSY814 (◆,▲, ■, respectively) by electroporation as well as Hyg3S/74NT-A into LSY678 (◆) and the correction efficiency was calculated as described above. The mean and standard deviation for each sample were determined from at least three independent experiments performed in duplicate and are represented in the table below the graph.
