Abstract
We previously demonstrated that a fraction of the human Nup107–160 nuclear pore subcomplex is recruited to kinetochores at the onset of mitosis. However, the molecular determinants for its kinetochore targeting and the functional significance of this localization were not investigated. Here, we show that the Nup107–160 complex interacts with CENP-F, but that CENP-F only moderately contributes to its targeting to kinetochores. In addition, we show that the recruitment of the Nup107–160 complex to kinetochores mainly depends on the Ndc80 complex. We further demonstrate that efficient depletion of the Nup107–160 complex from kinetochores, achieved either by combining siRNAs targeting several of its subunits excluding Seh1, or by depleting Seh1 alone, induces a mitotic delay. Further analysis of Seh1-depleted cells revealed impaired chromosome congression, reduced kinetochore tension and kinetochore–microtubule attachment defects. Finally, we show that the presence of the Nup107–160 complex at kinetochores is required for the recruitment of Crm1 and RanGAP1–RanBP2 to these structures. Together, our data thus provide the first molecular clues underlying the function of the human Nup107–160 complex at kinetochores.
Keywords: CENP-F, Crm1, Ndc80 complex, nucleoporin, Seh1
Introduction
Kinetochores are macromolecular complexes that assemble on centromeric chromatin during mitosis to ensure accurate segregation of sister chromatids (reviewed in Maiato et al, 2004; Chan et al, 2005). Over the last years, an increasing number of kinetochore constituents have been identified and insights into their contributions to kinetochore assembly and function have emerged. However, deciphering the composition, assembly and function of this key cell division structure remains an important challenge.
The inner plate of the mature vertebrate kinetochore contains constitutive centromere constituents that provide a structural platform for the recruitment of the components of the outer kinetochore layers, namely the outer plate and fibrous corona (Liu et al, 2006 and references therein). The outer kinetochore domain comprises checkpoint-signaling molecules (including Mad1/2, Bub1/3, BubR1 and Mps1) that prevent anaphase onset until all chromosomes have achieved proper bipolar attachment to the spindle microtubules (MTs) (for review see Cleveland et al, 2003; Karess, 2005). These proteins likely monitor MT attachments that are specified by CENP-E, CENP-F, CLASP and the conserved KNL-1/Mis12 complex/Ndc80 complex network (reviewed in Maiato et al, 2004; Kline-Smith et al, 2005). The other major function of the outer kinetochore domain indeed involves the formation and maintenance of a stable kinetochore–MT interaction. A key player in this process is the evolutionarily conserved Ndc80 complex, comprised of Ndc80 (or its human homologue, Hec1), Nuf2, Spc24 and Spc25 (reviewed in Kline-Smith et al, 2005). Defects in the Ndc80 complex affect kinetochore–MT attachment, chromosome congression and chromosome segregation in all systems studied so far (reviewed in Maiato et al, 2004; Kline-Smith et al, 2005; see also Cheeseman et al, 2006; DeLuca et al, 2006). Another protein involved in MT attachment is CENP-F (also known as mitosin) (Rattner et al, 1993; Zhu et al, 1995). Depletion of CENP-F in HeLa cells was shown to impair kinetochore assembly in a subset of cells, a phenotype that might be correlated with its early recruitment to nascent kinetochores in late G2 (Bomont et al, 2005; Yang et al, 2005). However, the majority of CENP-F-depleted cells exhibited a strong checkpoint-dependent mitotic delay, with reduced tension between kinetochores and decreased stability of kinetochore MTs (Bomont et al, 2005; Holt et al, 2005; Laoukili et al, 2005; Yang et al, 2005; Feng et al, 2006).
In metazoans, the establishment of the mitotic spindle apparatus further requires the disassembly of the nuclear envelope (NE) during the prophase–prometaphase transition. This step also implies the dispersal of the nuclear pore complexes (NPCs), which are macromolecular assemblies anchored within the NE that control nucleocytoplasmic transport during interphase. Over recent years, major progresses have been made concerning the molecular mechanisms underlying NE and NPC reassembly around the two newly formed nuclei in late anaphase and telophase (reviewed in Hetzer et al, 2005). Time-course studies initially revealed the sequential recruitment of the NPC constituents, the nucleoporins, to the reforming nucleus. In particular, the constituents of the Nup107–160 complex, which remain associated throughout mitosis, are among the earliest nucleoporins recruited on the chromatin surface in anaphase (Belgareh et al, 2001). This evolutionarily conserved complex, which is composed in vertebrates of nine different nucleoporins (Nup160, Nup133, Nup107, Nup96, Nup85, Nup43, Nup37, Sec13 and Seh1), is stably associated on both faces of the NPCs during interphase, suggesting a structural role of this complex within NPCs (Belgareh et al, 2001; Vasu et al, 2001). In addition, siRNA-mediated depletion of the Nup107–160 complex and in vitro approaches based on Xenopus egg extracts demonstrated a crucial role of the Nup107–160 complex at a very early stage of NPC assembly (Boehmer et al, 2003; Harel et al, 2003; Walther et al, 2003).
We previously reported that a fraction (∼5–10%) of the entire human Nup107–160 complex can be found at kinetochores from prophase—even before nuclear envelope breakdown—to late anaphase, when NPCs are already reforming on chromatin surface (Belgareh et al, 2001; Loiodice et al, 2004). In addition, this complex was also reported to be localized to spindle poles and proximal spindle fibers in prometaphase mammalian cells, and throughout reconstituted spindles in Xenopus egg extracts (Orjalo et al, 2006).
Besides this complex, an increasing number of proteins have now been demonstrated to share dual localization or function at both kinetochores and NPCs. This includes the checkpoint proteins Mad1, Mad2 and Mps1 and the mitotic checkpoint regulator Rae1 (reviewed in Stukenberg and Macara, 2003; see also Babu et al, 2003; Liu et al, 2003). In addition, the small GTPase Ran and transport receptors of the karyopherin/importin β family play a critical role in both mitotic spindle assembly and kinetochore function (Arnaoutov and Dasso, 2003; Harel and Forbes, 2004). In particular, the Ran-GTP binding nuclear export receptor (exportin) Crm1 was demonstrated to be localized at kinetochores, where it provides an anchoring site for RanGAP1 and RanBP2/Nup358 (Arnaoutov et al, 2005), two proteins that localize to the cytoplasmic side of NPCs in interphase (Matunis et al, 1998; Joseph et al, 2002, 2004). Finally, the putative transcription factor ELYS was recently shown to copurify with the Nup107–160 complex in vertebrates, and to localize to NPCs during interphase and to kinetochores throughout mitosis (Rasala et al, 2006).
Although depletion of Nup133 was recently reported to cause a delay in or failure to complete cytokinesis (Rasala et al, 2006), nothing was known so far concerning the molecular mechanisms underlying the targeting and function of the mammalian Nup107–160 complex at kinetochores. Here, we report that anchoring of the human Nup107–160 complex at kinetochores is mediated by the Ndc80 complex and CENP-F. In addition, we show that the Seh1 subunit of the Nup107–160 complex is essential for the targeting of this NPC subcomplex to kinetochores. We found that kinetochores depleted of Nup107–160 complex fail to establish proper MT attachments, thus inducing a checkpoint-dependent mitotic delay. Consistently, we found that the mitotic Ran-GTP effector, Crm1, as well as its binding partner, the RanGAP1–RanBP2 complex, are mislocalized upon depletion of the Nup107–160 complex from kinetochores.
Results
Nup133 interacts with the outer kinetochore protein CENP-F
In order to gain insights concerning the specific role of the Nup107–160 complex in mitosis, we used a yeast two-hybrid approach using human Nup133 as bait. This screen identified a C-terminal fragment of CENP-F (aa 2644–3065; CENP-FC) as a potential binding partner of Nup133. Interestingly, we independently identified a Nup133 peptide (IVFNAQGDSVLGAGACGGVPIIFSR, aa 442–467 of Nup133) in a systematic mass spectrometry analysis of CENP-F binding proteins, thereby strengthening the yeast two-hybrid result. Immunoprecipitation experiments indeed confirmed that CENP-F specifically co-precipitates with the Nup107–160 complex in colchicine-arrested extracts but not in asynchronous extracts (Figure 1A).
Figure 1.
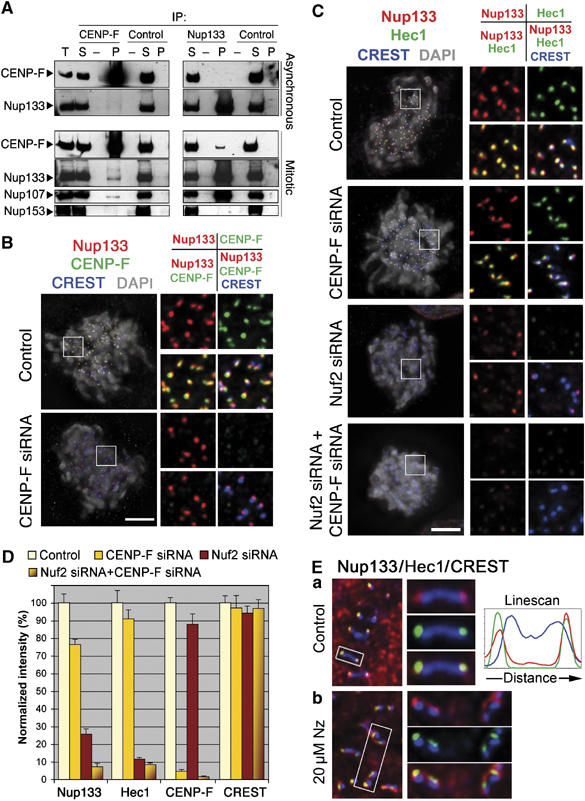
Nup107–160 complex targeting at kinetochores depends on CENP-F and the Ndc80 complex. (A) Immunoprecipitation of asynchronous or colchicine-arrested (mitotic) HeLa cell extracts using either anti-CENP-F or anti-myc (control) mouse IgG, or affinity-purified anti-hNup133 or anti-GST (control) rabbit IgG. Equivalent amounts of total extracts (T) and immune supernatants (S) and a 10-fold equivalent of the immune pellets (P) were analyzed by immunoblot using anti-CENP-F, anti-hNup133, anti-hNup107 and as control the mAb414 antibody that recognizes Nup153, a nucleoporin that does not belong to the Nup107–160 complex. Because of the lower amounts of CENP-F in asynchronous as compared with mitotic extracts, CENP-F immunoblots were exposed to yield comparable input signals. (B) HeLa cells treated for 3 days with control or CENP-F siRNA duplexes were pre-extracted, fixed and stained with anti-hNup133 (red), anti-CENP-F (green), CREST serum (blue) and DAPI (gray). (C) Cells treated for 3 days with control, CENP-F, Nuf2 or a combination of CENP-F and Nuf2 siRNA duplexes were processed for immunofluorescence as above using anti-hNup133 (red), anti-Hec1 (green), CREST serum (blue) and DAPI (gray). Maximum projection of deconvolved Z-stack images of all the four staining (left) or various combinations of overlay images (insets, right) is presented. Scale bar, 5 μm. (D) Quantification of the fluorescence intensity at kinetochores was performed as described in Materials and methods. (E) Single Z-plane of deconvolved immunofluorescence images from (a) control or (b) nocodazole-treated (Nz, 20 μM, 3 h) cells stained for Nup133 (red), Hec1 (green) or CREST antigens (blue). Right: enlargements of the marked areas stained with CREST and either Nup133 (top), Hec1 (center) or Nup133+Hec1 (bottom). Line scan through a single kinetochore pair, performed using the ‘Linescan' function accessed using the Metamorph software system, is also shown.
To characterize the biological relevance of this interaction, we analyzed the localization of Nup133 at kinetochores upon siRNA-mediated depletion of CENP-F. As shown in Figure 1B, a mild decrease of Nup133 signal at kinetochores could be detected in CENP-F-depleted cells. Quantification of fluorescence intensities at prometaphase kinetochores (see Materials and methods) indicated that the level of Nup133 was reduced by ∼25% as compared with cells transfected with a control siRNA (Figure 1D). In agreement with our previous data indicating that the Nup107–160 complex is targeted as one entity to kinetochores (Loiodice et al, 2004), a similar decrease was observed using specific anti-Nup107 antibodies (data not shown), or upon analysis of GFP-Seh1 in HeLa cells stably expressing this fusion (Supplementary Figure 1A and C). In contrast, and in agreement with previous studies (Bomont et al, 2005; Holt et al, 2005; Feng et al, 2006), CENP-F depletion did not significantly alter the targeting of Hec1 (Figure 1D). As CENP-F depletion was very efficient, this result suggested that CENP-F is only responsible for the localization of a minor fraction of the Nup107–160 complex to kinetochores.
CENP-F and the Ndc80 complex independently contribute to the recruitment of the Nup107–160 complex to the outer plate of kinetochores
To identify additional constituents involved in the targeting of the Nup107–160 complex to kinetochores, we analyzed the consequences of siRNA-induced depletion of other kinetochore proteins. Among the various siRNAs tested (see Supplementary Table I), those directed against Nuf2 or Hec1, which induce the depletion of the entire Ndc80 complex from kinetochores (DeLuca et al, 2002; Meraldi et al, 2004), led to a strong decrease of the Nup133 and GFP-Seh1 signals at kinetochores without affecting the soluble pool of the Nup107–160 complex (Figure 1C and D; Supplementary Figure 1B and C, and data not shown). In contrast, siRNA-induced depletion of CENP-E, Zwint-1, Zw10, Mad1, Mad2, Bub1, BubR1, RanGAP1, RanBP2 or INCENP had no effect on Nup133 (Supplementary Figure 2A and B, and data not shown). Therefore, the Ndc80 complex plays a major role in the recruitment of the Nup107–160 complex to kinetochores.
In agreement with these findings, fluorescence analyses revealed that in metaphase cells, Nup133 staining closely overlaps with the Hec1 antigen (Figure 1Ea). As recently reported (Orjalo et al, 2006), nocodazole treatment caused Nup133 to assemble as expanded crescents around the centromeres (Figure 1Eb; Supplementary Figure 3), a morphological change previously described for a subset of dynamic outer kinetochore proteins (Hoffman et al, 2001). A similar behavior was observed for CENP-F (Supplementary Figure 3), whereas the Ndc80 constituent Hec1 retained a more punctuate staining (DeLuca et al, 2005). Interestingly, this crescent localization of Nup133 in nocodazole-treated cells persisted upon depletion of Nuf2, but was sensitive to CENP-F depletion (Supplementary Figure 3). Finally, simultaneous depletion of both Nuf2 and CENP-F gave rise to an additive defect on the targeting of Nup133 and Nup107 to kinetochores of prometaphase cells (Figure 1C and D, and data not shown). Together, our data therefore indicate that the Nup107–160 complex is localized on the outer domain of kinetochores, where its anchoring mainly depends on the Ndc80 complex, whereas CENP-F may provide a more dynamic and MT-dependent binding site.
RNAi treatments that efficiently deplete the Nup107–160 complex at kinetochores induce a mitotic delay
Having established the molecular determinants within the kinetochores that specified the recruitment of the Nup107–160 complex, we aimed at testing the functional roles of this complex in mitosis. Therefore, we systematically analyzed the effect of siRNAs targeting various constituents of the Nup107–160 complex on its kinetochore localization. In agreement with a recent study (Rasala et al, 2006), siRNA-induced depletion of Nup133, while efficiently depleting the soluble pool of the protein, only moderately affected its kinetochore localization (Figure 2Aa). Consistently, quantification of Nup107 fluorescence intensity revealed that a significant fraction of Nup107 (∼50%) was still present at kinetochores upon depletion of Nup133 (Figure 2B and C). Similar results were obtained using siRNA against Nup160, Nup107, Nup85, Nup43 or Nup37 (data not shown). In contrast, depletion of Seh1 led to its efficient depletion including at kinetochores (Figure 2Ab and Supplementary Figure 4B) and significantly reduced the kinetochore levels of Nup107 (<10% residual staining) (Figure 2B and C), without affecting the stability of the other components of the complex (Figure 2D). Finally, combination of siRNAs targeting several Nup107–160 constituents (namely Nup160, Nup133, Nup107, Nup85, Nup43 and Nup37, but not Seh1, subsequently referred to as ‘combined siRNAs') led to a very efficient depletion of the entire complex, resulting in the lack of detectable staining at kinetochores, as revealed by anti-Nup107 and GFP-Seh1 staining (Figure 2B and C and Supplementary Figure 4B).
Figure 2.
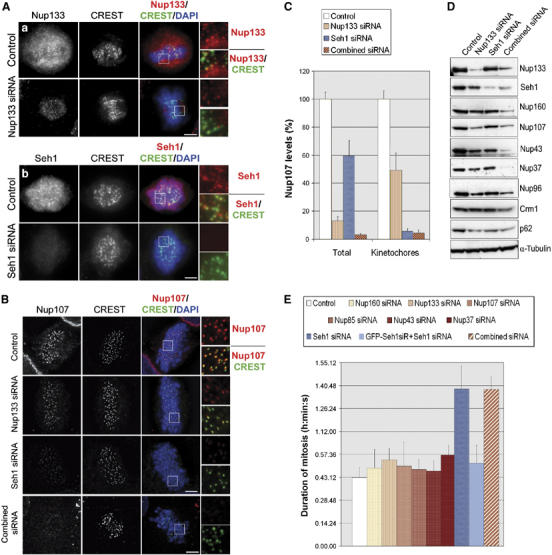
Seh1 RNAi, which prevents the Nup107–160 complex from localizing at kinetochores, or efficient depletion of the Nup107–160 complex, induces a mitotic delay. (A, B) HeLa cells transfected with the indicated siRNA duplexes were pre-extracted before fixation and stained with (Aa) anti-Nup133, (Ab) anti-Seh1 or (B) anti-Nup107 antibodies (red), CREST serum (green) and DAPI (blue). Wide-field microscopy images (A) or maximum projections of deconvolved Z-stacks (B) of representative prometaphase cells and three-fold enlargements of the marked area are shown. Scale bar, 5 μm. (C) Quantification of Nup107 fluorescence intensity in mitotic HeLa cells. Total levels were measured on wide-field images from non-extracted cells (n⩾20) and levels at kinetochores were measured on pre-extracted cells as in Figure 1D. (D) Whole-cell extracts from control, Nup133, Seh1 or ‘combined siRNA'-treated HeLa cells were prepared 3 days after transfection and analyzed by Western blot using the indicated antibodies. α-Tubulin is shown as loading control. (E) HeLa cells treated with the indicated siRNA duplexes were recorded by time-lapse videomicroscopy from 24 to 96 h post siRNA transfection, acquiring images every 5 or 10 min. Duration of mitosis was calculated as the time spent between the first frame where cells rounded up and the first frame where two distinct daughter cells could be observed. Average values of at least 50 cells for each condition are presented. For the complementation experiment (GFP-Seh1siR+Seh1 siRNA), a HeLa cell line stably expressing an siRNA-resistant form of GFP-Seh1 was used (see Supplementary Figure 4B).
We next used time-lapse microscopy to monitor the effect of these various siRNAs treatments on cell cycle progression. Whereas individually, siRNA-induced depletion of Nup133, Nup160, Nup107, Nup85, Nup43 or Nup37 only slightly affected mitotic progression, their combined depletion led to a significant increase of mitotic duration (from 45 min in control cells to more than 1 h and 30 min in the combined siRNA-treated cells; Figure 2E and Supplementary Figure 5). Of note, these cells died several hours after mitotic exit (Supplementary Figure 5), a phenotype that might be correlated with the drastic effect of the ‘combined siRNAs' on both nucleoporin stability and NPC assembly (Figure 2D and Supplementary Figure 4A). Importantly, Seh1 RNAi, which specifically prevented Nup107–160 complex recruitment to kinetochores (Figure 2B and C) without affecting its overall stability (Figure 2D), led to a similar increase of mitotic duration as the ‘combined siRNA' treatment (Figure 2E). The specificity of this phenotype was validated by the use of three distinct siRNA duplexes and by its complementation upon expression of an siRNA-resistant variant of GFP-Seh1 (Figure 2E and Supplementary Figure 4). As previously reported, Seh1 depletion had a minor effect on NPC assembly as compared with the depletion of other Nup107–160 constituents such as Nup133 or Nup107 (Loiodice et al, 2004). Accordingly, our data indicate that the mitotic delay observed upon siRNA-induced Seh1 depletion is unlikely to result from a global alteration of NPC assembly. As this mitotic defect was shared by the combined depletion of other constituents of the Nup107–160 complex, it appears to correlate with the efficient depletion of the Nup107–160 complex from kinetochores (see Discussion). We therefore focused subsequent analyses mainly on the phenotypes induced by Seh1 depletion, and confirmed the major phenotypes by using the ‘combined siRNA' treatment.
Seh1 depletion induces a checkpoint-dependent mitotic delay, impairs kinetochore–MT attachment and reduces tension at kinetochores of bi-oriented chromosomes
To further characterize this mitotic delay, we monitored mitotic progression in HeLa cells expressing histone H2B-GFP that were transfected with control or Seh1 siRNAs. As compared with control cells, Seh1-depleted cells displayed extended prometaphases associated with defects in chromosome congression and a delay in metaphase–anaphase transition (Figure 3A and B and Supplementary Movies 1, 2, 3, 4 and 5). Consistently, quantifications performed on cells fixed 3 days after siRNA treatment indeed revealed that misaligned chromosomes were present in more than 60% of Seh1-depleted cells in which a clear metaphase plate was observed, as compared with 5% in control cells (Figure 3C).
Figure 3.
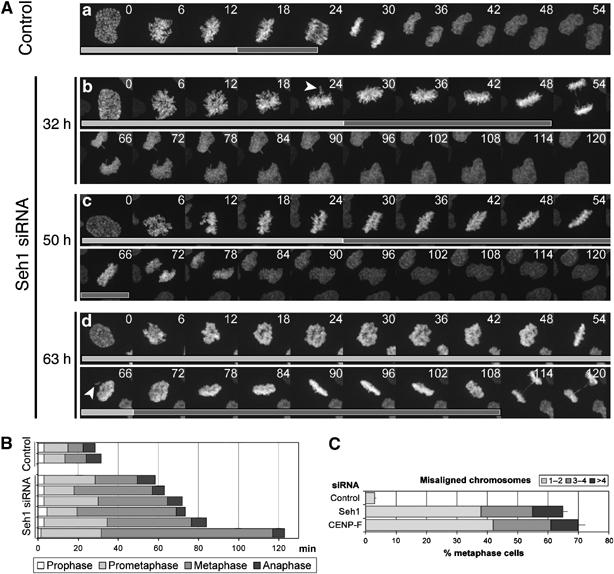
Seh1-depleted cells display extended prometaphase and metaphase. (A) HeLa cells transiently expressing histone H2B-GFP were treated with control (a) or Seh1 (b–d) siRNAs. At 32 h (b), 50 h (c) or 63 h (d) after siRNA transfection, randomly chosen prophase cells were imaged every 90 s for several hours with a spinning disk confocal microscope. Maximum intensity projections of 14 Z-stack planes for each time point are presented. Arrowheads indicate misaligned chromosomes. Light gray bars indicate frames from prophase to metaphase, whereas dark gray bars outline time spent from metaphase to anaphase. (B) Mitotic progression in two control and six representative Seh1-depleted cells selected at random. (C) Quantification of misaligned chromosomes (defined as in Meraldi and Sorger (2005), as chromosomes located outside a rectangular area encompassing the central 30% of the spindle) in control, Seh1- or CENP-F siRNA-treated cells. The number of cells displaying 1–2 (light gray), 3–4 (dark gray) or >4 (black) misaligned chromosomes is expressed as percentage of total metaphase cells analyzed (at least 100 per condition).
To determine whether these mitotic defects are monitored by the spindle assembly checkpoint, we analyzed the localization of Mad2, Mad1 and BubR1. Upon Seh1 depletion, Mad2 and Mad1 antibodies positively stained kinetochores from misaligned chromosomes, as well as a few kinetochores from the metaphase plates (Figure 4A and Supplementary Figure 6A). In addition, a persistent BubR1 signal was detected on all kinetochores (Figure 4B). In agreement with these observations, time-lapse videomicroscopy revealed that the combined depletion of Seh1 and either Mad2 or BubR1 by RNAi abrogates the Seh1-induced mitotic delay (Supplementary Figure 6B). Together, our data therefore indicate that Seh1 depletion induces defects in both chromosome congression and metaphase–anaphase transition, and that these defects are sensitive to the spindle assembly checkpoint.
Figure 4.
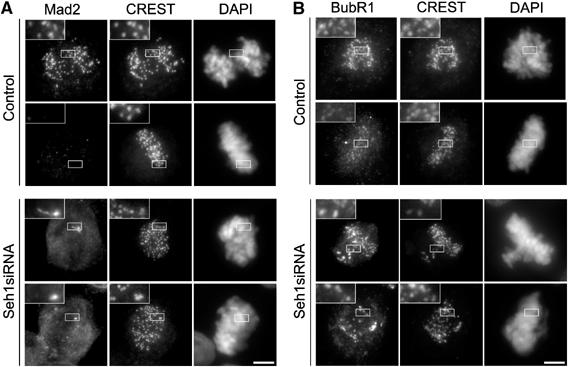
Seh1 depletion activates the spindle assembly checkpoint. Immunofluorescence of metaphase or prometaphase HeLa cells upon 72 h treatment with control or Seh1 siRNA stained for Mad2 (A) or BubR1 (B), CREST and DAPI. Three-fold magnifications of the marked areas are presented. Scale bar, 5 μm.
To better characterize these chromosome congression and segregation defects, we next examined the kinetochore–MT attachments. In Seh1 siRNA-treated cells, no detectable MTs were observed in proximity of kinetochores from misaligned chromosomes (Figure 5A, compare c and d). In addition, although most congressed chromosomes appeared to be attached to MTs when cells were fixed at 37°C (Figure 5Ba), fewer and less organized cold-stable kinetochore MTs (also referred to as K-fibers) were present in Seh1-depleted cells as compared with control cells (Figure 5Bb). In line with this observation, suggesting improper or unstable kinetochore–MT attachment (Rieder, 1981), spindles were generally much longer in Seh1-depleted cells at metaphase (Figure 5C). Finally, quantification of the interkinetochore spacing revealed that tension-mediated stretching between sister kinetochores of aligned chromosomes was reduced in Seh1-depleted cells (inter-kinetochore distance of 0.84±0.06 versus 1.68±0.11 μm for control cells; Figure 5D). In agreement with the persistence of some cold-stable kinetochore MTs, some tension remained however, as uncongressed kinetochores present in Seh1-depleted cells, or fully relaxed kinetochores measured in control cells after nocodazole-mediated MT disassembly, showed even smaller spacing (0.61±0.07 and 0.54±0.05 μm, respectively; Figure 5D).
Figure 5.
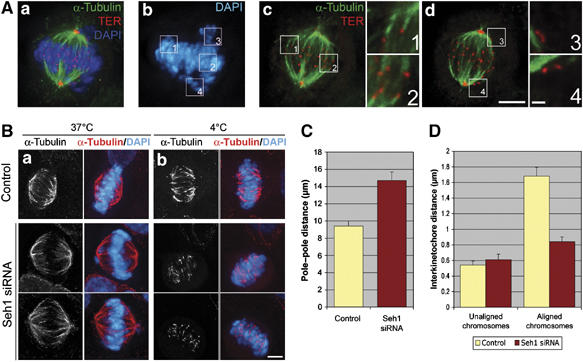
Seh1 depletion impairs chromosome congression and reduces tension between sister kinetochores. (A) Seh1-depleted cells were fixed and then stained with anti-α-tubulin (green), the TER serum that labels both the centromeres and centrosomes (red), and DAPI (blue). A maximum intensity projection of the deconvolved Z-stack from a representative mitotic cell is presented in (a) and (b). Single planes of the same cell illustrating kinetochore–microtubule attachment on chromosomes of the metaphase plate (c) and lack of microtubules on misaligned chromosomes (d). Scale bar, 5 μm. Higher magnifications of the marked areas are presented (scale bar, 1 μm). (B) Immunofluorescence analysis of control or Seh1-depleted cells placed either at 37°C (a) or at 4°C (b) for 10 min before fixation and stained with anti-α-tubulin antibody (red) and DAPI (blue). Scale bar, 5 μm. (C) Pole–pole distance was calculated as the distance between the two centrosomes labelled with the TER serum as in (A). (D) Interkinetochore distance between aligned or unaligned sister kinetochores situated in the same focal plane were calculated on deconvolved images. Values for unaligned chromosomes in control cells was obtained after treatment with 20 μM nocodazole for 3 h. At least 50 kinetochore pairs in eight different cells were measured.
Together, these observations indicate that Seh1-depleted cells display defects both in chromosome attachment and congression, and in the formation of stable MT–kinetochore interactions. Noteworthy, analysis of cells treated with the (combined siRNAs) similarly revealed an increased spindle length (Supplementary Figure 7A and B), suggesting that the phenotypes induced by Seh1 depletion are likely due to the depletion of the Nup107–160 complex from kinetochores.
The Nup107–160 complex acts upstream of RanGAP1–RanBP2 at kinetochores
To determine whether the targeting of the Nup107–160 complex and its anchoring determinants could be interdependent, we next investigated the localization of the Ndc80 constituent, Hec1, and of CENP-F in Seh1-depleted cells. Hec1 localization was not affected by this treatment, whereas a significant decrease of CENP-F levels was observed on congressed kinetochores of Seh1-depleted cells as compared with normal metaphase kinetochores (Figure 6A). However, kinetochores from unattached polar chromosomes were still CENP-F positives (Figure 6A) and nocodazole treatment of Seh1-depleted cells restored the localization of CENP-F at all kinetochores (Figure 6B). These data thus indicate that the presence of the Nup107–160 complex at kinetochores, although dispensable for the recruitment of CENP-F to these structures, is required for its maintenance at attached yet tension-defective kinetochores.
Figure 6.
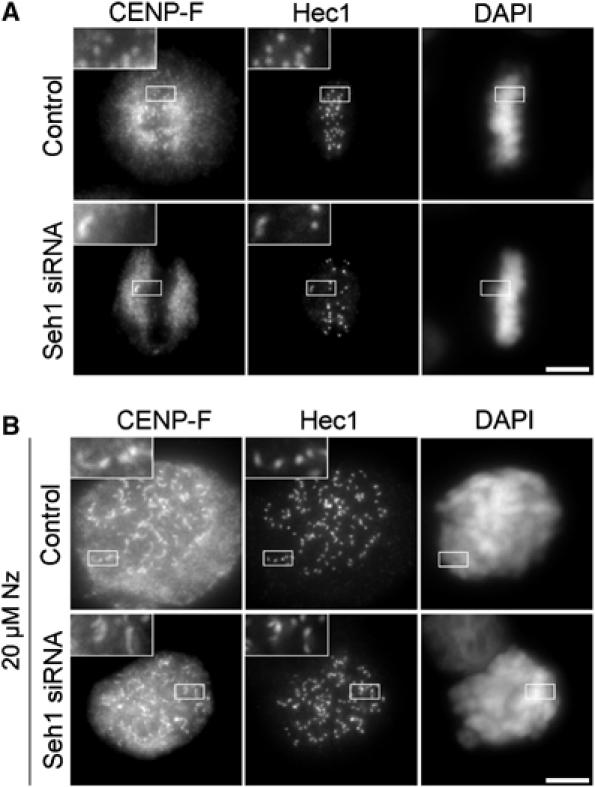
Seh1 depletion induces MT-dependent stripping of CENP-F. Cells transfected with control or Seh1 siRNAs were either fixed and stained with anti-Hec1 and anti-CENP-F antibodies and DAPI (A), or treated for 3 h with 20 μM nocodazole before fixation (B). Three-fold enlargements of the marked areas are presented. Scale bar, 5 μm.
It was previously reported that Hec1 and Nuf2 are essential for targeting both RanGAP1 and RanBP2 to kinetochores (Joseph et al, 2004). We thus investigated the localization of the RanGAP1–RanBP2 complex and of its anchoring determinant at kinetochore, Crm1 (Arnaoutov et al, 2005). Immunofluorescence analysis revealed a significant decrease of kinetochore levels of RanGAP1 (which reflects the localization of the RanGAP1–RanBP2 complex; Joseph et al, 2002) and of Crm1 in Nuf2- or Seh1-depleted cells, as well as in cells treated with the ‘combined siRNAs' (Figure 7A and B). Whereas the mislocalization of the RanGAP1–RanBP2 complex could be an indirect consequence of the defects in the MT–kinetochore attachment occurring in these cells, Crm1 binding to kinetochores was reported to be independent of MT attachment (Arnaoutov et al, 2005). Consistently, Crm1 was still mislocalized from kinetochores upon nocodazole treatment of Seh1-depleted cells (data not shown). Together, our results therefore indicate that treatments that prevent the localization of the Nup107–160 complex at kinetochores (Nuf2, Seh1 or ‘combined siRNAs') all impair the kinetochore targeting of Crm1, and thus of the ternary complex composed of Ran-GTP, Crm1 and its cargo, RanGAP1–RanBP2, to this structure.
Figure 7.
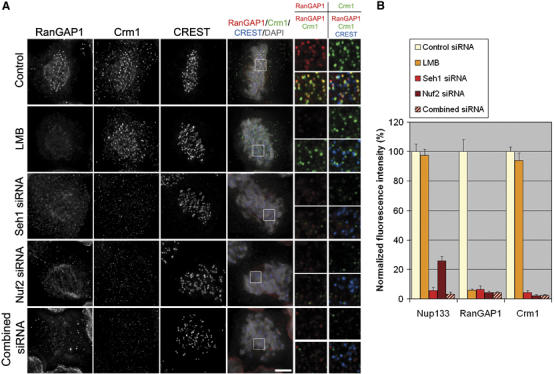
Mislocalization of the Nup107–160 complex from kinetochores impairs the kinetochore targeting of Crm1 and RanGAP1. (A) Cells were treated for 20 min with 10 ng/ml leptomycin B or for 72 h with control, Seh1, Nuf2 or the ‘combined siRNA' duplexes and subsequently pre-extracted, fixed and probed with anti-RanGAP1 (red) and anti-Crm1 (green) antibodies, CREST serum (blue) and DAPI. Single planes from deconvolved Z-stack images (scale bar, 5 μm) and three-fold enlargements of the boxed areas are presented. (B) Remaining levels of Nup133, RanGAP1 and Crm1 at kinetochores in control cells, upon treatment with Seh1, Nuf2 or the combined RNAi, or after LMB treatment, were quantified based on deconvolved Z-stack images (see Materials and methods).
Discussion
Anchoring of the Nup107–160 complex at the outer kinetochore domain
In this study, we have demonstrated that efficient targeting of the Nup107–160 complex to kinetochores requires the Ndc80 complex and CENP-F, both of which were previously localized within the kinetochore outer plate (Zhu et al, 1995; DeLuca et al, 2005). In agreement with the current view of the temporal regulation of the kinetochore assembly pathway, the Nup107–160 complex is, like both the Ndc80 complex and CENP-F, recruited to these structures in late G2/early prophase (Liao et al, 1995; Belgareh et al, 2001; Martin-Lluesma et al, 2002; Loiodice et al, 2004). However, both the Ndc80 and Nup107–160 complexes persist at kinetochores until late anaphase (Belgareh et al, 2001; Martin-Lluesma et al, 2002; Loiodice et al, 2004), whereas CENP-F relocalizes to the spindle midzone in early anaphase (Liao et al, 1995). At that stage, the Ndc80 complex likely becomes crucial for the maintenance of the Nup107–160 complex at kinetochores. While the link between CENP-F and the Nup107–160 complex could be confirmed by immunoprecipitation experiments performed on colchicine-arrested cells—a condition that likely favors their interaction as indicated by our immunofluorescence data (Supplementary Figure 3A)—we were not able so far to demonstrate a molecular interaction between the Ndc80 complex and any constituent of the Nup107–160 complex (our unpublished results). This suggests that this interaction is either biochemically unstable or indirect, that is, mediated by a so far uncharacterized partner of the Ndc80 complex. The dual targeting of the Nup107–160 complex to kinetochores could reflect a simultaneous dependence on its two anchoring determinants or the presence of two distinct pools of the Nup107–160 complex at kinetochores (Figure 8). However, our RNAi experiments performed on nocodazole-arrested cells as well as our previous FRAP and FLIP experiments (Belgareh et al, 2001) rather argue for the latter hypothesis. In either case, it is likely that the assembly of the Nup107–160 complex falls along the CENP-I assembly pathway, which was recently shown to be essential for recruiting CENP-F and the Ndc80 complex to the kinetochore through two separate assembly branches (Liu et al, 2006).
Figure 8.
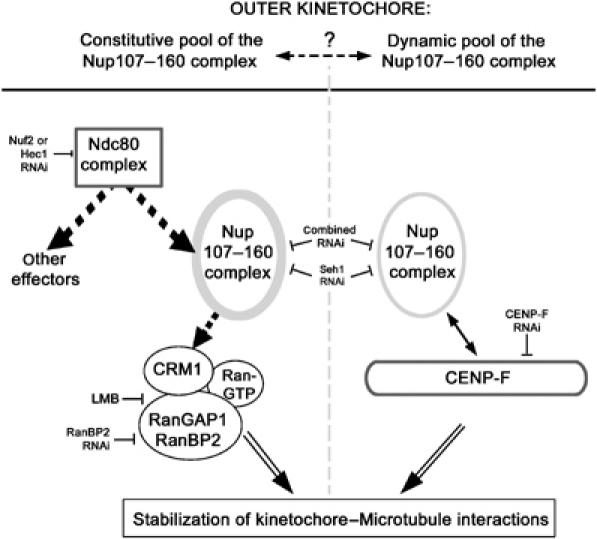
Model summarizing the protein network involved in the targeting and function of the Nup107–160 complex at kinetochores. Dotted arrows indicate direct or indirect interactions. As outlined in Discussion, Seh1 is comprised within the Nup107–160 complex in this model.
Seh1 is required for kinetochore localization of the Nup107–160 complex
Functional analysis of the human Nup107–160 complex in mitosis had so far been hampered by its crucial involvement in NPC assembly (Harel et al, 2003; Walther et al, 2003) and by the persistence of this complex at kinetochore upon RNAi-induced depletion of several of its constituents (namely Nup133, Nup107, Nup85, Nup43 or Nup37; Rasala et al, 2006 and our unpublished data; Figure 2). Here, we demonstrate that efficient depletion of the entire Nup107–160 complex from kinetochores can be achieved either by co-depleting several of its constituents, or by only depleting one of its subunits, Seh1. Both treatments led to a significant delay in mitosis, an increased spindle length in metaphase and impaired kinetochore localization of Crm1, thus revealing for the first time the biological relevance of the kinetochore localization of the Nup107–160 complex. Because Seh1 depletion does not significantly impair NPC assembly as compared with other Nup107–160 complex constituents, these mitotic defects are unlikely to simply result from a primary defect in NPC assembly. Although in HeLa cells Seh1 could not be efficiently co-precipitated with the other constituents of the human Nup107–160 complex (see however the discussion in Loiodice et al, 2004), our data indicate that GFP-Seh1 behaves as the other constituents of this complex upon various RNAi treatments, thereby strengthening its behavior as a Nup107–160 constituent. How Seh1 contributes to the kinetochore localization of the Nup107–160 complex remains to be elucidated. This protein could either provide a physical link between the Nup107–160 complex and its anchoring sites at kinetochores, or be required to turn this complex in a conformation or a particular status suitable for its kinetochore localization and mitotic function.
Of note, efficient depletion of the Nup107–160 complex upon our ‘combined siRNAs' treatment affects the stability of the whole complex, including Seh1 or Nup96 that are not directly targeted by this combined RNAi treatment (see Figure 2D). Accordingly, the mitotic phenotypes associated with the efficient depletion of the Nup107–160 complex in mitosis may reflect the function of either the whole complex or one of its loosely associated constituents, Seh1. In particular, we cannot exclude that Seh1 might have additional Nup107–160-independent functions in mitosis. In the future, characterization of additional factors interacting with the Nup107–160 complex, and more specifically with Seh1, will thus be required to better understand the respective contribution of Seh1 versus the other constituents of the Nup107–160 complex in proper kinetochore function.
Downstream effectors of the Nup107–160 complex at kinetochores
It was previously suggested that RanBP2 recruitment to kinetochores may be an important aspect of Hec1 and Nuf2 function (Joseph et al, 2004). Our study revealed that depletion of either Nuf2 or Seh1 or the ‘combined siRNAs' treatment, all of which lead to the efficient depletion of the Nup107–160 complex from kinetochores, also impair the kinetochore localization of Crm1 and the RanGAP1–RanBP2 complex. Consistently, these various treatments as well as depletion of RanBP2 lead to an increased spindle length, a phenotype proposed to be the consequence of decreased tension at kinetochores (DeLuca et al, 2002; Salina et al, 2003; Joseph et al, 2004; McCleland et al, 2004). Of note, depletion of the Nup107–160 complex from kinetochores does not entirely recapitulate the phenotypes of the Ndc80 complex, a result likely reflecting the direct or indirect role of the Ndc80 complex in mediating kinetochore–MT attachment (reviewed in Kotwaliwale and Biggins, 2006).
Although further studies will be required to characterize at the molecular level the (direct or indirect) interactions between Ndc80, the Nup107–160 complex and the Crm1/RanGTP/RanGAP1–RanBP2 complex, our data indicate that the Nup107–160 complex behaves as a downstream effector of the Ndc80 complex, required for the localization of Crm1 at kinetochores throughout mitosis (Figure 8). In addition, our results suggest that Seh1 is also required to stabilize kinetochore-bound CENP-F from MT-dependent stripping. As defects in chromosome congression and tension were also reported upon CENP-F depletion (Bomont et al, 2005; Holt et al, 2005; Yang et al, 2005; Feng et al, 2006), CENP-F may also contribute to some of the phenotypes occurring upon Seh1 depletion and/or kinetochore mislocalization of the Nup107–160 complex (Figure 8). In mitosis, the Nup107–160 complex may thus contribute to integrate distinct kinetochore functions required for proper MT–kinetochore interactions that depend on separate assembly pathways.
Multiple functions of the vertebrate Nup107–160 complex in mitosis
Past studies demonstrated a critical role for the Nup107–160 complex in NPCs reformation upon mitotic exit both in Xenopus extracts and in mammalian cells (Boehmer et al, 2003; Harel et al, 2003; Walther et al, 2003). Recently, the Nup107–160 complex was also shown to be required for correct bipolar assembly in Xenopus extracts (Orjalo et al, 2006). Now our study indicates that, in HeLa cells, efficient depletion of the Nup107–160 complex, achieved by simultaneous depletion of several of its constituents, affects progression through mitosis, but has no major effect on spindle assembly (see Supplementary Figure 7). This discrepancy may be related to the fact that the Nup107–160 complex is broadly localized to spindle and Ran-induced asters in Xenopus extracts (Orjalo et al, 2006), whereas in HeLa cells it mainly localizes at kinetochores and can only be transiently observed at spindle poles and proximal spindle fibers (Loiodice et al, 2004; Orjalo et al, 2006). Moreover, whereas some of the defects associated with the depletion of the Nup107–160 complex from kinetochores in human cells can be explained by the subsequent loss of Crm1 from kinetochores, neither Crm1 nor the RanGAP1–RanBP2 complex could be found so far at kinetochores in Xenopus extracts (Arnaoutov and Dasso, 2005). As previously discussed for Crm1 (Arnaoutov and Dasso, 2005), these differences in localization and function of the Nup107–160 complex may potentially reflect differing mechanisms of spindle assembly in HeLa cells versus Xenopus egg extracts.
In agreement with our videomicroscopy experiments, Rasala et al (2006) reported that partial loss of Nup133 from kinetochores did not result in clear spindle assembly or mitotic chromosome alignment defects. Yet, they found that depletion of Nup133 causes a delay in or failure to complete cytokinesis, a phenotype that might be related to our previous observation, namely the accumulation of G1 cells characterized by cytoplasmic SC-35 foci (Loiodice et al, 2004). Whether this cytokinesis defect is an indirect consequence of a primary function of the Nup107–160 complex in NPC assembly or chromosome segregation, or corresponds to a novel function of this complex in mitosis remains to be determined.
Conclusion
Our work has revealed the molecular mechanisms underlying the anchoring of the Nup107–160 complex to kinetochores, together with its requirement for chromosome congression and segregation. Whether the dual localization and function of this complex at NPCs and kinetochores helps to coordinate entry, progression and exit from mitosis will be a challenging question for future research.
Materials and methods
Yeast two-hybrid, immunoprecipitation and immunoaffinity purification of CENP-F binding proteins
The pLex12-hNup133 construct (Belgareh et al, 2001) was used as bait to screen a random primed human placenta cDNA library cloned into the pP6 plasmid, using a mating approach as described (Formstecher et al, 2005).
HeLa and HeLa S3 cells were cultured and synchronized as described (Loiodice et al, 2004). Immunoprecipitation experiments were performed essentially as described (Loiodice et al, 2004) using affinity-purified anti-Nup133, anti-GST, anti-CENP-F (clone 11; BD Biosciences) or anti-myc (clone 9E10; Sigma-Aldrich, St Louis, MO) antibodies.
For immunoaffinity purification of CENP-F binding proteins, a HeLa nuclear extract eluted from a phosphocellulose column with 0.5 M KCl and dialyzed into 0.25 M KCl/20 mM Tris–HCl pH 7.5/0.5% NP-40 was passed over a CENP-F antibody column. The column was washed with 0.5 M KCl and eluted with 0.2 M glycine pH 2.2. Mass spectrometric peptide sequencing was performed as described (Bochar et al, 2000).
siRNA transfection, immunofluorescence microscopy, deconvolution and fluorescence quantification at kinetochores
Sequences of the siRNA duplexes used in this study are provided in Supplementary Table I. siRNAs were transfected using either Oligofectamine (Invitrogen) as previously described (Loiodice et al, 2004) or HiPerfect (Qiagen) (with 10 nM siRNA), both of which led to similar depletion efficiency. For the ‘combined siRNAs' treatment, 10 nM of each siRNA was used. For all control experiments, siRNAs targeting GFP or Lamin A/C were used.
To establish HeLa cell lines stably expressing an siRNA-resistant form of GFP-Seh1, the GFP-Seh1si(a)R fusion was generated by site-directed mutagenesis using the Quickchange kit (Stratagene) (TAG → CTC at positions 165–167) and then inserted into the pIRES-neo vector (Clontech). HeLa cells were transfected using the calcium phosphate procedure and individual clones were isolated by G418 selection.
Antibodies used in this study are listed in Supplementary data. For immunofluorescence, cells were either fixed in 3% paraformaldehyde (PAF) and subsequently permeabilized with 0.5% Triton X-100, as described (Loiodice et al, 2004), or washed in a solution containing 4% PAF+0.05% glutaraldheyde, pre-extracted for 2 min in 0.5% Triton X-100 and then fixed for 20 min in 3% PAF. Wide-field microscopic images (Z-stacks) were acquired and deconvolved as previously described (Loiodice et al, 2004).
Quantifications of fluorescence intensities at kinetochores were carried out on deconvolved 3D images. Kinetochores were identified using the multidimensional image analysis spot detection software based on wavelet transform segmentation (developed in Institut Curie as a Metamorph module), and described in 3D by their gravity center for a reference marker. Fluorescence intensities for all proteins were then calculated in 2D in circular regions of 8 pixels (0.5 μm) of diameter around the gravity centers of the reference kinetochore marker. The value of 100% was assigned to the fluorescence intensity of each marker in control cells. For each condition, at least 100 kinetochores belonging to five different cells were quantified and their intensity was averaged. For quantification of RNAi experiments, only mitotic cells whose kinetochores were efficiently depleted (as assessed by double labelling) were chosen for analysis.
Live cell microscopy
HeLa cells were seeded onto 35-mm glass dishes (Iwaki) and kept in open chambers equilibrated in 5% CO2 and maintained at 37°C. For phase-contrast imaging, time-lapse sequences were taken every 5 or 10 min for 72 h using a × 20 objective on a Leica DMIRBE inverted microscope equipped with a cooled CCD camera (Micro Max 5 MHz; Ropper Scientific) and controlled by the Metamorph software (Universal Imaging). For GFP-H2B imaging, HeLa cells were transfected by the calcium phosphate procedure with pBOS-H2B-GFP (BD PharMingen) 16 h before siRNA transfection. The medium was exchanged with prewarmed Opti-MEM medium containing 10% of fetal bovine serum and 0.2 μg/ml vitamin C sodium salt 2 h before recording. Imaging was conducted on a Perkin-Elmer UltraView RS Nipkow-Disk system attached to a Zeiss Axiovert 200M microscope equipped with a × 63/1.4NA objective. Stacks of 14 images every 1.5 μm were collected every 90 s with a Cooled CCD Hamamatsu ORCA II ER camera.
Supplementary Material
Supplementary Figures
Supplementary Table I
Supplementary Information
Supplementary Movies Legends
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Movie 4
Supplementary Movie 5
Acknowledgments
We are grateful to V Cordes, Ed Salmon, M Bornens and R Karess for providing antibodies, to M Yoshida for Leptomycin B, J Camonis and F Perez (BioPhenics Project—Translational Department, Institut Curie) for providing certain siRNAs, to the Curie Imaging, the Plate-Forme d'Imagerie Dynamique of Institut Pasteur and Hybrigenics Staffs and members of ours laboratories for constant support and valuable comments during the course of this work.This work was supported by the Institut Curie, the Centre National de la Recherche Scientifique, the Ligue contre le Cancer (comité de Paris), the Ligue Nationale Contre le Cancer (équipe labellisée 2006), the Association pour la Recherche contre le Cancer and the Ministère de l'Education Nationale, de la Recherche et de l'Enseignement Supérieur (A.C.I. ‘Jeunes chercheurs') (to VD), a GenHomme Network Grant (02490-6088) to Hybrigenics and Institut Curie, by Grants from the Ministry of Education, Science, Sports and Culture of Japan (MEXT) to TF by the fondation pour la recherche médicale (to VG), by NIH (CA099423, CA75138 and core grant CA06927), and an Appropriation from the Commonwealth of Pennsylvania to TJY. MZ was supported by scholarship from the Ecole Normale Supérieure and from Boehringer Ingelheim Fonds and AA and VR by fellowships of the Ministère délégué à la Recherche et aux nouvelles Technologies.
References
- Arnaoutov A, Azuma Y, Ribbeck K, Joseph J, Boyarchuk Y, Karpova T, McNally J, Dasso M (2005) Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat Cell Biol 7: 626–632 [DOI] [PubMed] [Google Scholar]
- Arnaoutov A, Dasso M (2003) The Ran GTPase regulates kinetochore function. Dev Cell 5: 99–111 [DOI] [PubMed] [Google Scholar]
- Arnaoutov A, Dasso M (2005) Ran-GTP regulates kinetochore attachment in somatic cells. Cell Cycle 4: 1161–1165 [DOI] [PubMed] [Google Scholar]
- Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, Van Deursen JM (2003) Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol 160: 341–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N, Rabut G, Bai SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont-Racine M, Ellenberg J, Doye V (2001) An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol 154: 1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochar DA, Wang L, Beniya H, Kinev A, Xue Y, Lane WS, Wang W, Kashanchi F, Shiekhattar R (2000) BRCA1 is associated with a human SWI/SNF-related complex: linking chromatin remodeling to breast cancer. Cell 102: 257–265 [DOI] [PubMed] [Google Scholar]
- Boehmer T, Enninga J, Dales S, Blobel G, Zhong H (2003) Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc Natl Acad Sci USA 100: 981–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomont P, Maddox P, Shah JV, Desai AB, Cleveland DW (2005) Unstable microtubule capture at kinetochores depleted of the centromere-associated protein CENP-F. EMBO J 24: 3927–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan GK, Liu ST, Yen TJ (2005) Kinetochore structure and function. Trends Cell Biol 15: 589–598 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A (2006) The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127: 983–997 [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112: 407–421 [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, McEwen BF (2005) Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell 16: 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED (2006) Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127: 969–982 [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED (2002) hNuf2 inhibition blocks stable kinetochore–microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol 159: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Huang H, Yen TJ (2006) CENP-F is a novel microtubule-binding protein that is essential for kinetochore attachments and affects the duration of the mitotic checkpoint delay. Chromosoma 115: 320–329 [DOI] [PubMed] [Google Scholar]
- Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, Jacq B, Arpin M, Bellaiche Y, Bellusci S, Benaroch P, Bornens M, Chanet R, Chavrier P, Delattre O, Doye V, Fehon R, Faye G, Galli T, Girault JA, Goud B, de Gunzburg J, Johannes L, Junier MP, Mirouse V, Mukherjee A, Papadopoulo D, Perez F, Plessis A, Rosse C, Saule S, Stoppa-Lyonnet D, Vincent A, White M, Legrain P, Wojcik J, Camonis J, Daviet L (2005) Protein interaction mapping: a Drosophila case study. Genome Res 15: 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A, Forbes DJ (2004) Importin beta: conducting a much larger cellular symphony. Mol Cell 16: 319–330 [DOI] [PubMed] [Google Scholar]
- Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ (2003) Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell 11: 853–864 [DOI] [PubMed] [Google Scholar]
- Hetzer MW, Walther TC, Mattaj IW (2005) Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol 21: 347–380 [DOI] [PubMed] [Google Scholar]
- Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED (2001) Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell 12: 1995–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SV, Vergnolle MA, Hussein D, Wozniak MJ, Allan VJ, Taylor SS (2005) Silencing Cenp-F weakens centromeric cohesion, prevents chromosome alignment and activates the spindle checkpoint. J Cell Sci 118: 4889–4900 [DOI] [PubMed] [Google Scholar]
- Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M (2004) The RanGAP1–RanBP2 complex is essential for microtubule–kinetochore interactions in vivo. Curr Biol 14: 611–617 [DOI] [PubMed] [Google Scholar]
- Joseph J, Tan S, Karpova T, McNally J, Dasso M (2002) SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J Cell Biol 156: 595–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess R (2005) Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol 15: 386–392 [DOI] [PubMed] [Google Scholar]
- Kline-Smith SL, Sandall S, Desai A (2005) Kinetochore–spindle microtubule interactions during mitosis. Curr Opin Cell Biol 17: 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwaliwale C, Biggins S (2006) Microtubule capture: a concerted effort. Cell 127: 1105–1108 [DOI] [PubMed] [Google Scholar]
- Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH (2005) FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol 7: 126–136 [DOI] [PubMed] [Google Scholar]
- Liao H, Winkfein RJ, Mack G, Rattner JB, Yen TJ (1995) CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J Cell Biol 130: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ST, Chan GK, Hittle JC, Fujii G, Lees E, Yen TJ (2003) Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol Biol Cell 14: 1638–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ST, Rattner JB, Jablonski SA, Yen TJ (2006) Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol 175: 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, Doye V (2004) The entire Nup107–160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell 15: 3333–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, DeLuca J, Salmon ED, Earnshaw WC (2004) The dynamic kinetochore–microtubule interface. J Cell Sci 117: 5461–5477 [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma S, Stucke VM, Nigg EA (2002) Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297: 2267–2270 [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Wu JA, Blobel G (1998) SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol 140: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland ML, Kallio MJ, Barrett-Wilt GA, Kestner CA, Shabanowitz J, Hunt DF, Gorbsky GJ, Stukenberg PT (2004) The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore–microtubule attachment. Curr Biol 14: 131–137 [DOI] [PubMed] [Google Scholar]
- Meraldi P, Draviam VM, Sorger PK (2004) Timing and checkpoints in the regulation of mitotic progression. Dev Cell 7: 45–60 [DOI] [PubMed] [Google Scholar]
- Meraldi P, Sorger PK (2005) A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J 24: 1621–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjalo AV, Arnaoutov A, Shen Z, Boyarchuk Y, Zeitlin SG, Fontoura B, Briggs S, Dasso M, Forbes DJ (2006) The Nup107–160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell 17: 3806–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ (2006) ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci USA 103: 17801–17806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner JB, Rao A, Fritzler MJ, Valencia DW, Yen TJ (1993) CENP-F is a ca 400 kDa kinetochore protein that exhibits a cell-cycle dependent localization. Cell Motil Cytoskeleton 26: 214–226 [DOI] [PubMed] [Google Scholar]
- Rieder CL (1981) The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma 84: 145–158 [DOI] [PubMed] [Google Scholar]
- Salina D, Enarson P, Rattner JB, Burke B (2003) Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J Cell Biol 162: 991–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenberg PT, Macara IG (2003) The kinetochore NUPtials. Nat Cell Biol 5: 945–947 [DOI] [PubMed] [Google Scholar]
- Vasu S, Shah S, Orjalo A, Park M, Fischer WH, Forbes DJ (2001) Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J Cell Biol 155: 339–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T, Mattaj IW, Doye V (2003) The conserved Nup107–160 complex is critical for nuclear pore complex assembly. Cell 113: 195–206 [DOI] [PubMed] [Google Scholar]
- Yang Z, Guo J, Chen Q, Ding C, Du J, Zhu X (2005) Silencing mitosin induces misaligned chromosomes, premature chromosome decondensation before anaphase onset, and mitotic cell death. Mol Cell Biol 25: 4062–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Chang KH, He D, Mancini MA, Brinkley WR, Lee WH (1995) The C terminus of mitosin is essential for its nuclear localization, centromere/kinetochore targeting, and dimerization. J Biol Chem 270: 19545–19550 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Table I
Supplementary Information
Supplementary Movies Legends
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Movie 4
Supplementary Movie 5


