Abstract
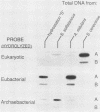
A procedure was developed for harvesting gram quantities of microbial biomass from oligotrophic waters, when mixed populations are present in low abundance. Picoplankton from Atlantic Ocean (Hydrostation S, Sargasso Sea) and Pacific Ocean (Aloha Station) sites were collected in a three-stage process: (i) collection of seawater through an intake covered with 10-microns-pore Nytex; (ii) concentration by a tangential flow filtration device equipped with 10 ft2 (0.929 m2) of 0.1-micron-pore fluorocarbon membrane; (iii) collection of cells from concentrate by centrifugation. The overall efficiency of picoplankton recovery was at least 37%. The cellular morphotypes recovered matched those of the original population. DNA was prepared from frozen cell pellets by enzymatic digestion, solvent extraction, and isopycnic centrifugation. As indicated by the binding of kingdom-specific hybridization probes to the purified DNA, the Sargasso Sea picoplankton in this collection were largely eubacteria.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ferguson R. L., Buckley E. N., Palumbo A. V. Response of marine bacterioplankton to differential filtration and confinement. Appl Environ Microbiol. 1984 Jan;47(1):49–55. doi: 10.1128/aem.47.1.49-55.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman J. A., Comeau D. E., Hagström A., Chan A. M. Extraction from natural planktonic microorganisms of DNA suitable for molecular biological studies. Appl Environ Microbiol. 1988 Jun;54(6):1426–1429. doi: 10.1128/aem.54.6.1426-1429.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S. J., DeLong E. F., Olsen G. J., Pace N. R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988 Feb;170(2):720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie J. E., Daley R. J., Jasper S. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977 May;33(5):1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogure K., Simidu U., Taga N. A tentative direct microscopic method for counting living marine bacteria. Can J Microbiol. 1979 Mar;25(3):415–420. doi: 10.1139/m79-063. [DOI] [PubMed] [Google Scholar]
- Lee S., Fuhrman J. A. Relationships between Biovolume and Biomass of Naturally Derived Marine Bacterioplankton. Appl Environ Microbiol. 1987 Jun;53(6):1298–1303. doi: 10.1128/aem.53.6.1298-1303.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., Lane D. J., Giovannoni S. J., Pace N. R., Stahl D. A. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- Sgaramella V., Khorana H. G. CXII. Total synthesis of the structural gene for an alanine transfer RNA from yeast. Enzymic joining of the chemically synthesized polydeoxynucleotides to form the DNA duplex representing nucleotide sequence 1 to 20. J Mol Biol. 1972 Dec 28;72(2):427–444. doi: 10.1016/0022-2836(72)90155-6. [DOI] [PubMed] [Google Scholar]
- Somerville C. C., Knight I. T., Straube W. L., Colwell R. R. Simple, rapid method for direct isolation of nucleic acids from aquatic environments. Appl Environ Microbiol. 1989 Mar;55(3):548–554. doi: 10.1128/aem.55.3.548-554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson S. W., Novitsky T. J., Quinby H. L., Valois F. W. Determination of bacterial number and biomass in the marine environment. Appl Environ Microbiol. 1977 Apr;33(4):940–946. doi: 10.1128/aem.33.4.940-946.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]