Figure 2.

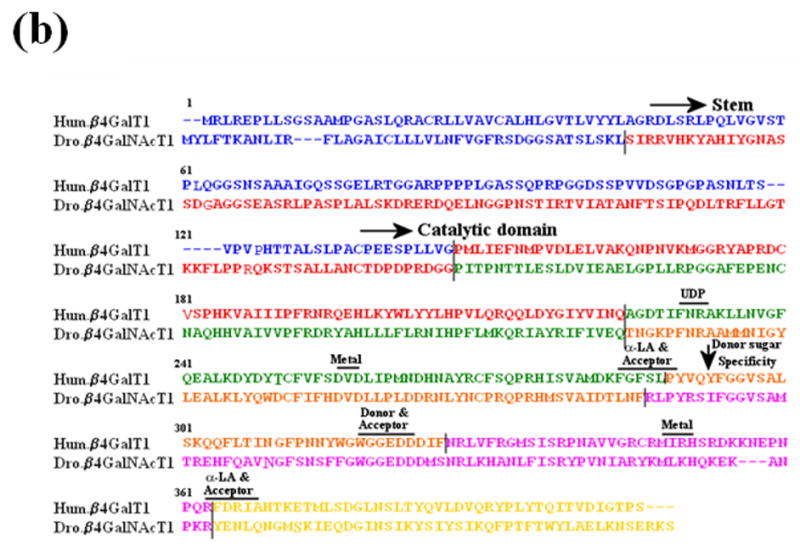
The sequence comparison of a region of vertebrate and invertebrate ortholog genes of β1,4Gal-T. (a) The sequence comparison of the region containing Tyr286 in human β1,4Gal-T1 with the corresponding region in the ortholog proteins of vertebrates (I) and invertebrates (II). In some β1,4Gal-T homologs in vertebrates, Phe is substituted for the residue Tyr at position 286. (b) The sequence comparison of the functional regions of human-β1,4Gal-T1 with Drosophila CG8536 (Dro.) protein. The two proteins show about 49% overall similarity and about 56% similarity in the catalytic domain. Ile at position 289 of Dro. protein (arrow), which corresponds to residue Tyr at position 289 or 286 in bovine or human β1,4-Gal-T1, respectively, that imparts the β4GalNAc-T1 activity in the Dro. protein. The residues involved in the binding of metal ion, acceptor, sugar donor, and α-LA,16, 8 are conserved in the two proteins. The sequence regions with different colors represent the regions coded by corresponding exons, and the line represents the intron-exon junctions.
