Figure 1.
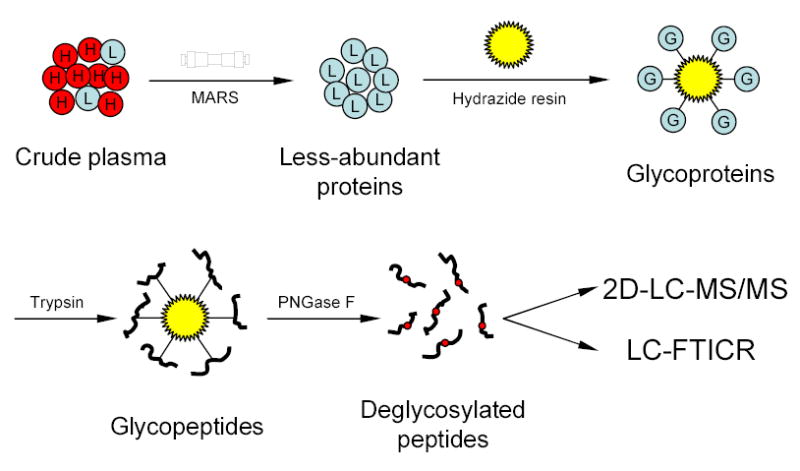
Strategy for plasma N-glycoprotein analysis using immunoaffinity subtraction, glycoprotein capture, and mass spectrometry. Crude plasma (red balls marked with “H” represent high abundance proteins; blue balls marked with “L” represent low abundance proteins) is first subjected to multi-component depletion using the pre-packed MARS column, followed by incubating the flow-through (minus the retained abundant proteins) with hydrazide resin. The captured glycoproteins (blue balls marked with “G”) are then washed and digested on-resin with trypsin, and the N-glycopeptides are specifically released from the resin by PNGase F. The resulting deglycosylated peptides can either be identified by 2-D LC-MS/MS or directly analyzed by LC-FTICR to validate the number of glycosylation sites.
