Fig. 1.
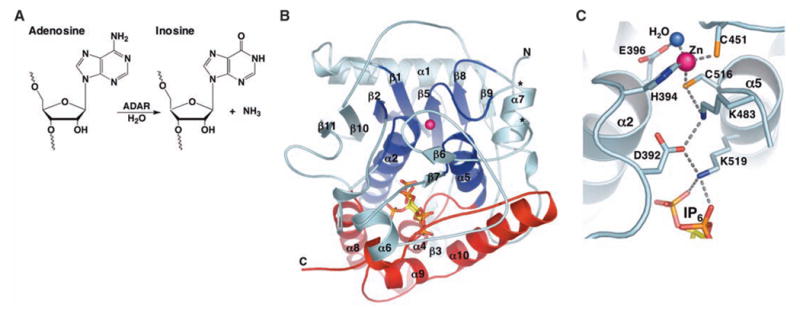
(A) ADAR catalyzed hydrolytic deamination of adenosine to inosine in dsRNA. (B) Ribbon model of hADAR2-D. The active-site zinc atom is represented by a magenta sphere. The N-terminal α/β domain (residues 306 to 620) is colored cyan, with the region that shares structural similarity with CDA and TadA colored dark blue (deamination motif; residues 350 to 375, 392 to 416, 439 to 455, 514 to 525, and 542 to 551). The C-terminal helical domain (residues 621 to 700), which with contributions from the deamination motif makes the major contacts to IP6 (ball and stick), is colored red. Ends of the disordered segment (residues 462 to 473) are indicated with asterisks. (C) Residue interactions at the active site. Shown are the zinc ion, coordinating residues (H394, C451, and C516), the nucleophilic water (blue sphere), and the proposed proton-shuttling residue, E396. The hydrogen-bond relay that connects the active site to the IP6 is also indicated. Single-letter abbreviations for amino acid residues are defined in (42).
