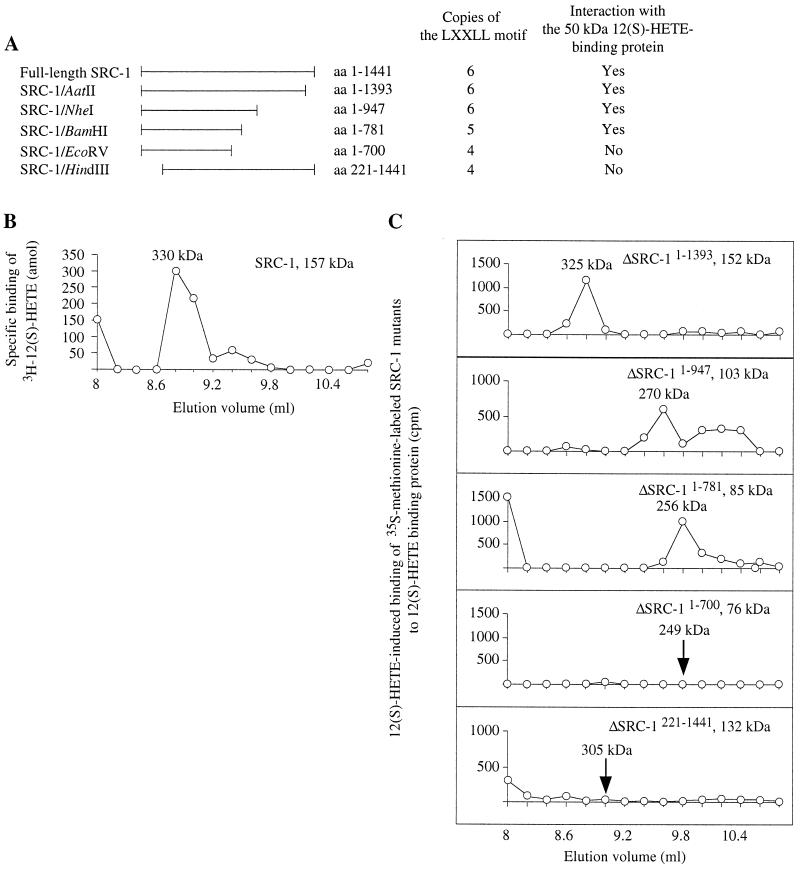
Figure 4.
Mapping of SRC-1 interaction domains for 50-kDa 12(S)-HETE binding protein. (A) Preparation and properties of SRC-1 deletion mutants (for details see Material and Methods). (B) Cytosol from LLC cells was treated with 10 mM ATP and chromatographed on Superdex 200. The fraction containing the 50-kDa 12(S)-HETE binding protein was incubated with 2 nM 12(S)-[3H]HETE for 1 h before the addition of unlabeled SRC-1. This incubation was allowed to proceed additionally for1 h at 4°C. The sample was rechromatographed on Superdex 200, and fractions were collected for radioactivity measurements. (C) Interaction assays. Cytosol from LLC cells was treated with 10 mM ATP and chromatographed on Superdex 200 to isolate the 50-kDa 12(S)-HETE binding protein. This protein was incubated with or without 1 nM 12(S)-HETE for 30 min at 4°C before addition of [35S]methionine-labeled deletion mutants of SRC-1 (30 min at 4°C). Samples were analyzed by gel permeation chromatography on Superdex 200, and radioactivity was measured in the fractions collected. The chromatograms show radioactivity in 12(S)-HETE-incubated fractions minus radioactivity in fractions from control experiments without ligand.