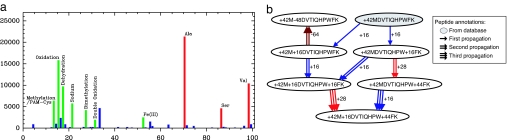
Fig. 2.
Discovery of modifications by using spectral networks. (a) Histogram of absolute parent mass differences for all detected spectral pairs on the IKKβ data set; the y axis represents the number of spectral pairs with a given difference in parent mass. For clarity, we only show the mass range 1–100 Da. The peaks at masses 71, 87, and 99 Da correspond to amino acid masses, and the peaks at masses 14, 16, 18, 22, 28, 32, and 53 Da correspond to known modifications that were also found by Tsur et al. (7) using blind database search. The peak at mass 34 Da corresponds to a putative modification that remains unexplained to date. (b) Modification network for peptide MDVTIQHPWFK from the Lens data set. The shaded node was annotated as peptide + 42MDVTIQHPWFK by database search of the tag VTIQHP; the remaining nodes were annotated by iterative propagation. On each propagation, the source peptide annotation is combined with the modification determined by the spectral product to yield a new peptide annotation (different modifications are shown as edges with different colors).