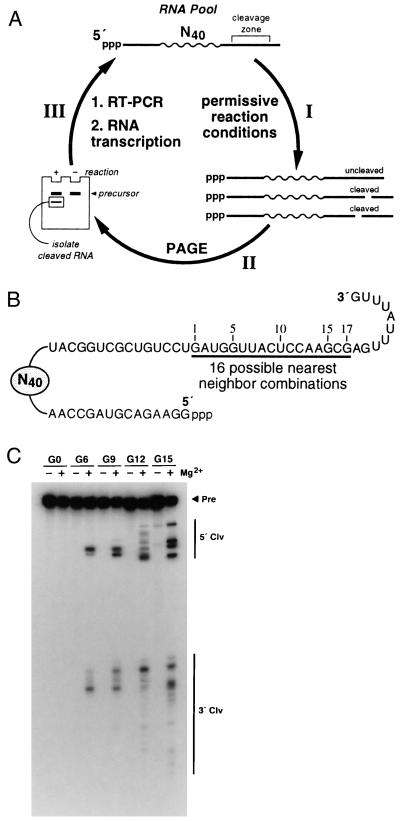
Figure 1.
In vitro selection of self-cleaving ribozymes. (A) Selection of self-cleaving ribozymes from random-sequence RNA. (I) RNAs are incubated under permissive reaction conditions (see text). (II) The 5′ fragments of cleaved RNAs are separated from uncleaved precursors by PAGE and (III) are amplified by RT-PCR, which introduces a T7 promoter sequence and restores the nucleotides that were lost upon ribozyme cleavage. The resulting double-stranded DNAs are transcribed in vitro using T7 RNAP to generate the subsequent population of RNAs. (B) RNA construct used to initiate the in vitro selection process. N40 depicts 40 random-sequence nts. Underlined nucleotides (1–17) identify the region that represents all 16 possible nearest neighbor combinations. (C) An autoradiogram of a denaturing 10% PAGE revealing the self-cleavage activities of the starting RNA population (G0) and the populations at G6, G9, G12, and G15 after incubation in the absence (−) or presence (+) of 20 mM Mg2+ under otherwise permissive reaction conditions. The locations of the precursor (Pre), 5′ cleavage (5′ Clv), and 3′ cleavage (3′ Clv) fragments are indicated.