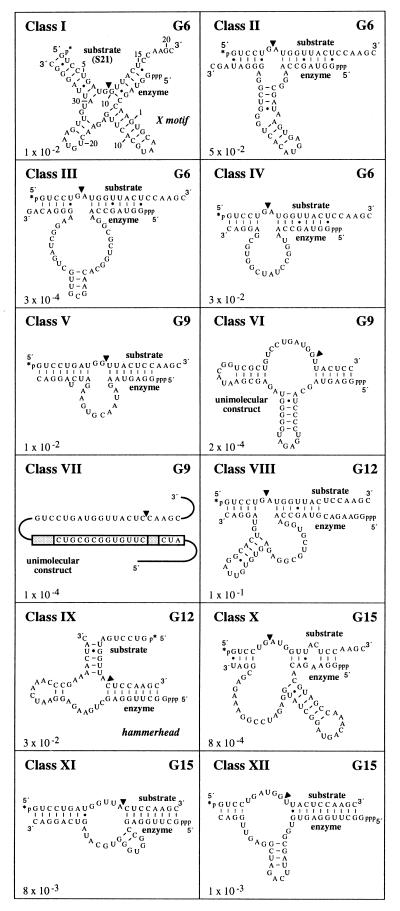
Figure 2.
Secondary structure models for 12 classes of self-cleaving ribozymes. Noted for each class is the generation at its first appearance, the rate constant for RNA cleavage (min−1), and the cleavage site (arrowhead). Secondary structure models are based on artificial phylogenetic data (e.g., see Fig. 3A) or otherwise reflect the most stable structure predicted by the Zuker RNA mfold program that is accessible on the internet (44). Putative Watson–Crick base pairing and G⋅U wobble interactions are represented by dashes and dots, respectively. For most classes, the predicted secondary structure was used as a guide to generate bimolecular ribozyme–substrate complexes that are truncated relative to the original clones. Classes VI and VII were examined as unimolecular constructs because preliminary efforts to generate bimolecular constructs were unsuccessful. For class VII, the boxed regions identify variable (shaded) or conserved nts that reside in the original random-sequence domain.