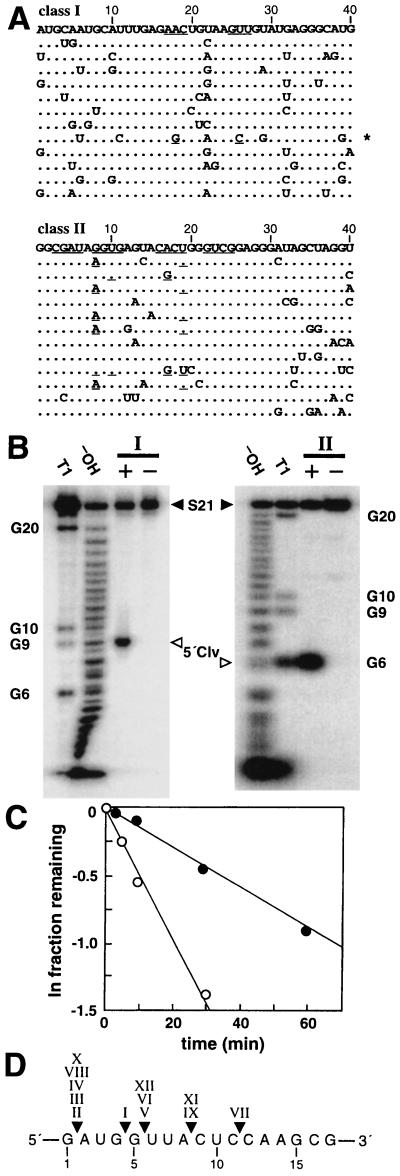
Figure 3.
Characterization of class I and class II self-cleaving ribozymes. (A) Sequence variations for artificial phylogenetic analysis are depicted for the nts corresponding to the original random-sequence domain (numbered). Dots indicate no change from the prototype (listed first). Underlined bases retain base complementary. For example, the asterisk identifies a class I ribozyme variant that carries mutations at positions 18 and 26 that retain the ability to base pair. (B) Determination of cleavage sites for bimolecular (Fig. 2) class I and class II ribozymes (Left and Right, respectively). In each case, a trace amount of 21-nt substrate (5′ 32P-labeled S21) was incubated with (+) or without (−) 500 nM ribozyme under the permissive reaction conditions. The 5′ cleavage fragments (5′ Clv) were separated from uncleaved S21 by denaturing 20% PAGE, and their sizes were compared with S21 5′ cleavage fragments prepared by partial ribonuclease (T1) and partial alkaline (−OH) digests. Class I and class II ribozymes cleave S21 after the G9 and G6 nts, respectively. (C) Cleavage reaction profiles for bimolecular class I (filled circles) and class II (open circles) ribozymes (Fig. 2) under the permissive reaction conditions. Observed rate constants for class I and class II ribozymes are 0.01 and 0.05 min−1, respectively, as determined by the negative slope of the lines representing the progression of each reaction. (D) Compilation of the cleavage sites of the 12 classes of ribozymes. Numbers identify the nts within the nearest neighbor domain depicted in Fig. 1B.