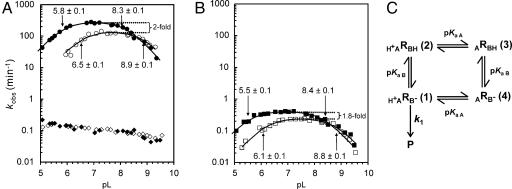
Fig. 1.
The effect of pH and D2O on cleavage rate. (A) Rate vs. pL (pH or pD) profiles of RG in H2O (filled circles), RG in D2O (open circles), RGkV in H2O (filled diamonds), and RGkV in D2O (open diamonds). (B) pL (pH or pD) profiles of 756C in H2O (filled squares) and in D2O (open squares). See Materials and Methods and Fig. 5 for a description of RNAs and mutants. The apparent first-order rate constant for cleavage (kobs) is plotted as a function of pL at 37°C (see Materials and Methods). (C) Kinetic model for self-cleavage of an RNA, R, which contains a single general acid of the functional form HA+ and single general base of the functional form B−. The extent of protonation of the acid and base is determined by their pKa values: pKaA and pKaB, respectively (from ref. 18). This model assumes the protonation states of the acid and base do not influence each other and that products (P) are only formed from RNA molecules in which both the general acid and base are in their active forms, H+ARB− (1); the proportion of RNA in this active form is abbreviated as fR(1). k1 is the intrinsic cleavage rate constant of the bond breaking step. Apparent pKa values for RG and 756C RNAs were estimated by fitting data to the equation kobs = fR(1) × k1 = k1/[1 + 10(pKaB−pH) + 10(pKaB−pKaA) + 10(pH−pKaA)] (18).