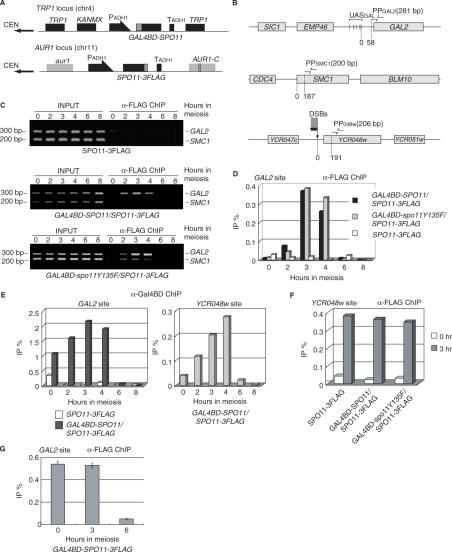
Figure 2.
Gal4BD–Spo11-dependent meiotic association of Spo11-3FLAG with the GAL2 UAS region in vivo. (A) SPO11-3FLAG was integrated into the AUR1 locus so that aureobasidin A could be used as a selection marker. GAL4BD-SPO11 (YHS425) or -spo11Y135F (YHS518) transgenes were integrated into the TRP1 locus. Both transgenes were constitutively expressed under the control of the ADH1 promoter (thick arrows) during the course of meiosis. Gray and black boxes represent the indicated loci. (B) Maps of the regions used for PCR detection in chromatin immunoprecipitation (ChIP). Five UAS sequences (indicated by vertical bars) exist around the GAL2 promoter region. PPGAL2 (indicated by the two arrows in the upper diagram) are located 58 bp away from the GAL2 locus. PPSMC1 (indicated by the two arrows in the middle diagram) amplifies an intragenic region of SMC1 (187 bp away from the BLM10 locus). PP048w (indicated by the two arrows in the lower diagram) is located 191 bp distal to the DSB site of the YCR048w locus. Gray boxes represent the indicated loci. (C) Meiotic binding of Spo11-3FLAG in the GAL2 UAS region in the presence (middle and lower panels) or absence (upper panel) of Gal4BD-fused protein (middle, Gal4BD-Spo11; lower, Gal4BD-spo11Y135F). Cells were crosslinked at 0, 2, 3, 4, 6 or 8 h of meiosis, and harvested to prepare WCEs from the cells expressing Spo11-3FLAG (YHS395), Gal4BD–Spo11/Spo11-3FLAG (YHS425) and Gal4BD–spo11Y135F/Spo11-3FLAG (YHS518). ChIP experiments were conducted using the WCEs and anti-FLAG antibody as described in the Materials and methods section. The immunoprecipitated (IP) DNA and the total genomic DNA from the WCEs (INPUT) were then amplified by 30 cycles of PCR and separated on a 3% agarose gel by electrophoresis. The upper and lower bands corresponding to the DNA fragment amplified by PPGAL2 (the GAL2 UAS region) and PPSMC1 (an internal control) are shown. The numbers at the left and upper sides of the panels indicate the positions of size markers (200 and 300 bp) and hours in SPM (lanes 0, 2, 3, 4, 6 and 8). (D) Quantitative real-time PCR (qPCR) of the immunoprecipitated DNA from samples taken from each time point as described in (C). The graph shows the kinetics of Spo11-3FLAG binding to the GAL2 UAS region during meiosis. The vertical axis indicates IP efficiencies (IP%) for the GAL2 UAS region, normalized with reference to the SMC1 locus. Numbers beneath the graphs are culture time (hours) in SPM. The experiment was performed independently twice. (E) Quantification of immunoprecipitated DNA at the GAL2 (left graph) and innate YCR048w (right graph) DSB sites by using the anti-Gal4BD antibody. The left graph shows the kinetics of Gal4BD–Spo11 binding to the GAL2 UAS site in YHS425 (co-expression of Gal4BD–Spo11 and Spo11-3FLAG) and YHS395 (expression of only Spo11-3FLAG). The right graph shows the ChIP signal at the innate YCR048w DSB site in YHS425. The experiment was performed independently twice. (F) Quantification of the anti-FLAG antibody-immunoprecipitated DNA at the YCR048w DSB sites in cells expressing Spo11-3FLAG alone, Gal4BD–Spo11 plus Spo11-3FLAG or Gal4BD-spo11Y135F plus Spo11-3FLAG. Open and gray bars represent signals at 0 and 3 h of meiosis. The ratios were normalized with reference to the values at the SMC1 locus. (G) Quantification of immunoprecipitated DNA using anti-FLAG antibody at the GAL2 in cells expressing Gal4BD–Spo11-3FLAG (YHS900). Numbers beneath the graphs are culture time (0, 3, 6 h) in SPM. The experiment was performed independently three times.