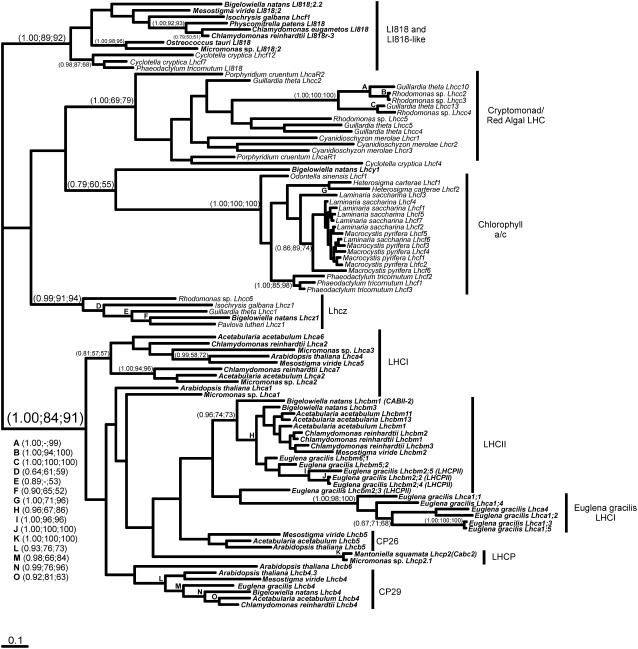
Figure 1.
Phylogenetic reconstruction of the LHC superfamily. The analysis includes sequences from several major light-harvesting divisions, including LI818 and LI818-like proteins, Chl a/c-binding proteins of the chromalveolates, the cryptomonad/red algal LHCs, the cryptophyte/haptophyte LHCs, and the Chl a/b-binding proteins. A MrBayes tree is shown (−lnL= 13,545.54, α = 1.419) with the support values for all analyses shown at specific nodes in the following order: MrBayes (posterior probabilities), NJD, PHYML. A total of 129 characters were included and the proportion of invariable sites was 0.041. The average sd of the split frequencies was 0.0126. A total of 100 sequences were included in the analyses and all sequences were either novel sequences generated in conjunction with the Protist EST Program (Table II), were present in GenBank, or were retrieved from individual genome projects (C. merolae). P. patens LI818 sequence compiled from individual ESTs available in dbEST (BJ958734, BJ957315, BJ955694, BJ954266, BJ958980, and BJ857081). Sequences from Chl a/b-containing organisms are in bold.