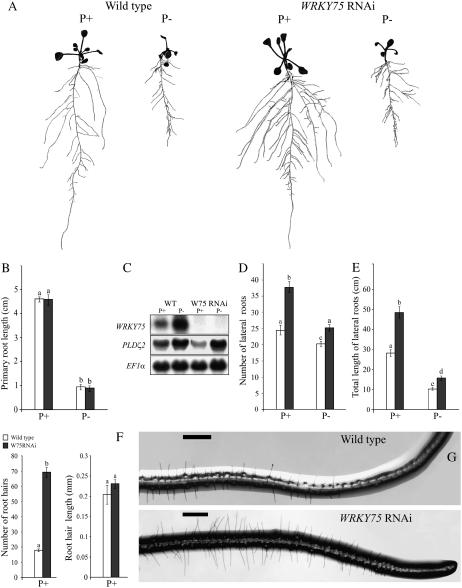
Figure 6.
Root architecture of WRKY75 RNAi plants. Wild-type and WRKY75 RNAi plants were grown under Pi-sufficient (P+) and Pi-deficient (P−) conditions for 7 d on vertically oriented petri plates. A, Lateral roots were spread to reveal the architectural details and scanned at 600 dpi. The seedlings shown are representative of 20 seedlings of the wild type and WRKY75 RNAi mutants grown under P+ and P− conditions. B, D, E, and F show comparative histograms of wild-type (white bars) and WRKY75 RNAi plants (black bars) with regard to various components of their root architecture under P+ or P− conditions. Different letters on the bars represent means that are statistically different (P < 0.02). Values are means ± se (n = 16) of each genotype per treatment. B, Primary root length. C, RNA-blot analysis showing the effect of WRKY75 RNAi knockdown on the expression of PLDζ2. Total RNA (15 μg) from wild-type (WT) and WRKY75 RNAi (W75 RNAi) plants grown under P+ and P− conditions was electrophoretically separated and blotted onto a nylon membrane. The membrane was initially hybridized with a 32P-labeled WRKY75 probe and later rehybridized with a probe for PLDζ2. EF1α was used as the loading control. D, Total length of all first-order lateral roots per plant. E, Total number of lateral roots per plant. F, Root hair length and number of root hair under P+ conditions. The data were recorded in an area spanning 5 mm from the root tip. Values are means ± se (n = 5) per genotype. G, Microscopic images of root tips with intact root hair from plants grown under P+ conditions. The scale bars in the panels represent 0.5 mm.