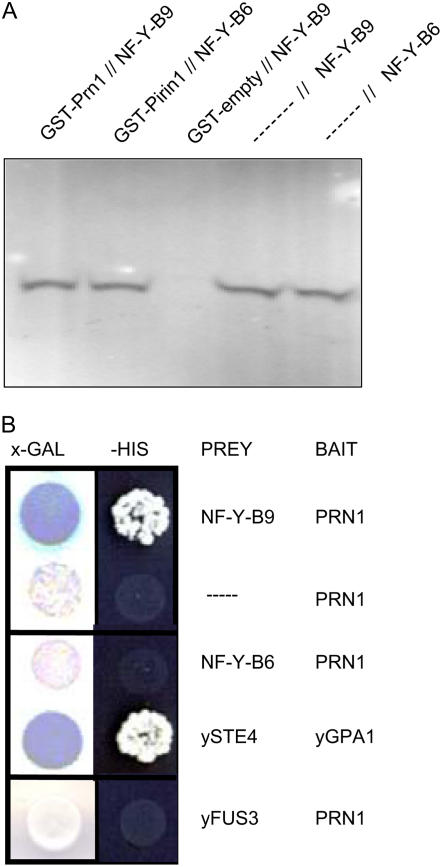
Figure 6.
PRN1 interacts with both NF-Y-B9 and NF-Y-B6. A, In vitro protein association assays. Full-length PRN1–GST fusions and radiolabeled translated NF-Y-B9 or NF-Y-B6 were coincubated in varying combinations. The PRN1-GST fusion, along with bound proteins were washed and resolved on 4% to 20% gradient SDS-PAGE gels as described (Lapik and Kaufman, 2003). Results were visualized using a Phosphor-Imager (Lapik and Kaufman, 2003). Lane 1: Full-length PRN1 interaction with full-length NF-Y-B9. Lane 2: Full-length PRN1 interaction with full-length NF-Y-B6. Lane 3: Negative control GST empty plasmid expressed protein does not interact with full-length NF-Y-B9 (LEC1). Lane 4: Full-length NF-Y-B9 alone (approximately 24 kD) as a molecular mass marker. Lane 5: Full-length NF-Y-B6 alone (approximately 24 kD) as a molecular mass marker. B, Yeast two-hybrid experiments. The potential for interaction between PRN1 and NF-Y-B9 or the closely related NF-Y-B6 was tested in a yeast two-hybrid assay (Lapik and Kaufman, 2003). The indicated combinations of the bait (pGBT9) and prey (pGAD424) constructs were transformed into the yeast reporter strain AH109. Empty vector (pGAD424) and the yeast mating response protein, yFUS3, were used as negative bait controls as shown. The interaction between the yeast Gα (yGPA1) and Gβ (ySTE4) was used as a positive interaction control. Initial transformants were plated on 9-cm sd plates (Trp−, Leu−, Lys−, His−) to select for His prototrophy. Positive transformants were also streaked on X-α-Gal-containing selective media where LacZ reporter gene activity was monitored visually. Blue color indicates positive interaction.