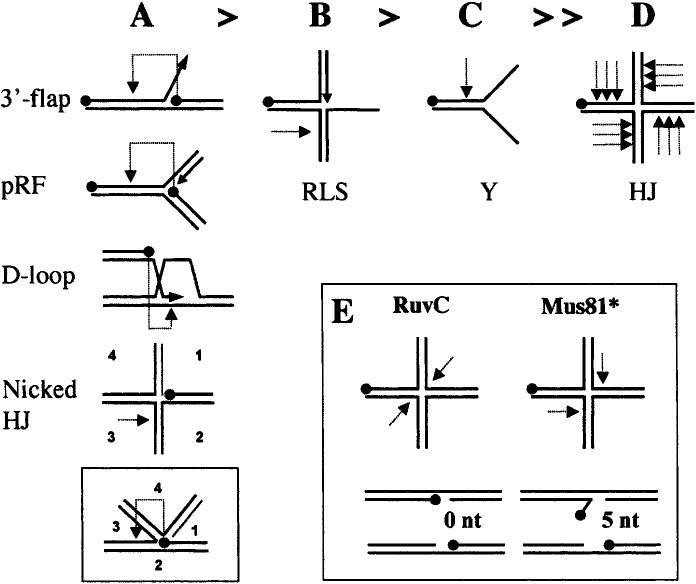
Figure 3.
Substrate-specificity of Mus81* in vitro. (A–D) Summary of results from a variety of cleavage studies using rMus81* with substrates presented in order of cleavage efficiency. The most efficient substrates (A) are cleaved well owing to the presence of a 5′-end at their branch junctions (indicated by a black dot). Cleavage of these substrates (light dotted arrow) is directed one-half helical turn upstream of this 5′-end (pRF, pseudoreplication fork). (A, inset) This mechanism is consistent with the cleavage of nicked HJs in the strand opposing the nick. Efficient cleavage of the remaining substrates (B–D) requires an excess of enzyme over substrate. A regressed lagging-strand substrate (RLS, panel B) is cleaved in the opposing strand, while a Y substrate (C) is cleaved upstream of the branch junction. A further increase in enzyme concentration results in nicking of mobile HJs at multiple sites on the 5′-side of the mobile core. (E) Contrast of the HJ cleavage products obtained by symmetrical cleavage with RuvC (nicked duplexes) to the unligatable flap and gap products produced by Mus81*. These 5′-flap products may isomerize into 3′-flaps that are further processed by Mus81* to produce only gapped molecules.