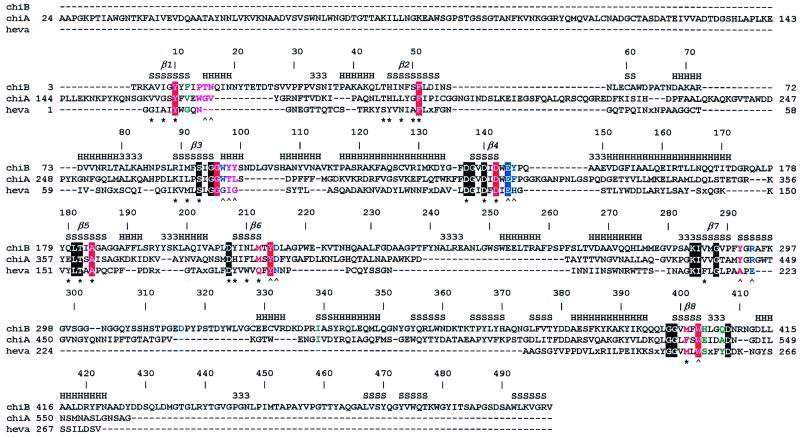
Figure 1.
Sequence alignment of chitinases. ChiB, chitinase B from S. marcescens; ChiA, chitinase A from S. marcescens; heva, hevamine from Hevea brasiliensis. An x in the sequences indicates a small insertion in hevamine. The secondary structure of ChiB is indicated at the top of the sequence alignment by H, α-helix; S, β-strand; 3, 310-helix. The TIM barrel β-strands are indicated by β1–8. Every 10th ChiB residue is labeled with its corresponding sequence number. Boxes indicate conserved residues. The color coding indicates the subsites of the chitotetraose model (see also Fig. 3A). Purple, −3; green, −2; red, −1; blue, +1. A ∧ indicates hydrogen bonding to an atom on the chitotetraose model, and a ★ indicates involvement of the side chain in the hydrogen-bonding network inside the TIM barrel core of ChiB. The catalytic Glu144 is part of the family 18 DxxDxDxE motif. The following definitions of loops and domains are used throughout the text: the porch loop (residues 14–27), the support loop (residues 233–262), the flexible loop (residues 315–325), the α/β-domain (residues 295–373) and the linker (residues 425–450), and the ChBD (residues 451–498).