Figure 1.
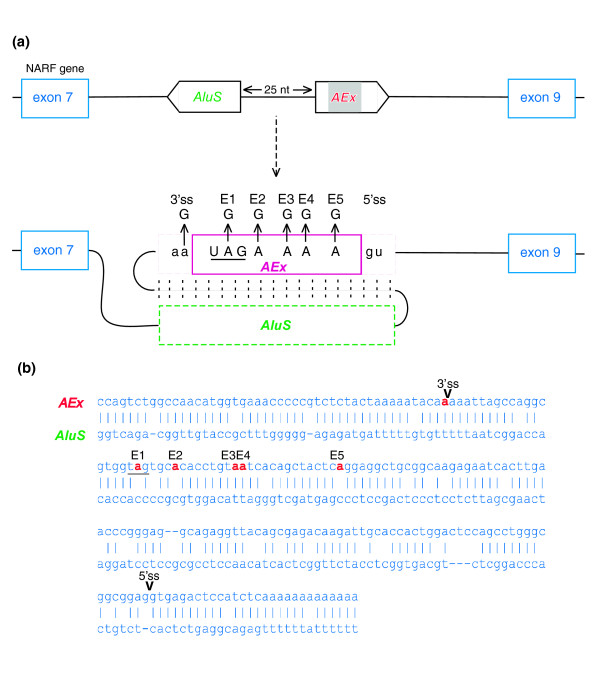
The birth of an Alu-exon through RNA editing. Editing prediction was inferred from alignment of cDNAs to human genomic DNA. (a) Schematic illustration of exons 7 to 9 of the NARF gene. Exons are depicted as blue boxes; the Alu-exon, derived from AluSx (AEx; purple box), is in a sense orientation and is shown in the middle. The intronic, antisense-orientation Alu sequence (AluS) is 25 base-pairs upstream of the exonized Alu. Sense and antisense Alus are expected to create a dsRNA secondary structure, thus allowing RNA editing. RNA editing changes an AA dinucleotide into a functional AG 3' splice site (lower panel). RNA editing also occurs in five positions in the Alu-exon itself (E1, E2, E3, E4 and E5). In the first position (E1), editing changes a UAG stop codon into a UGG Trp codon. (b) Predicted folding between the sense and antisense Alu sequences (upper and lower lines, respectively). Adenosines that undergo editing are marked by red. Splice sites utilized for Alu exonization are marked as 5' ss and 3' ss on the alignment.
