Figure 4. Brn3a binding to the Pou4f1 locus in vitro and in embryonic trigeminal ganglia.
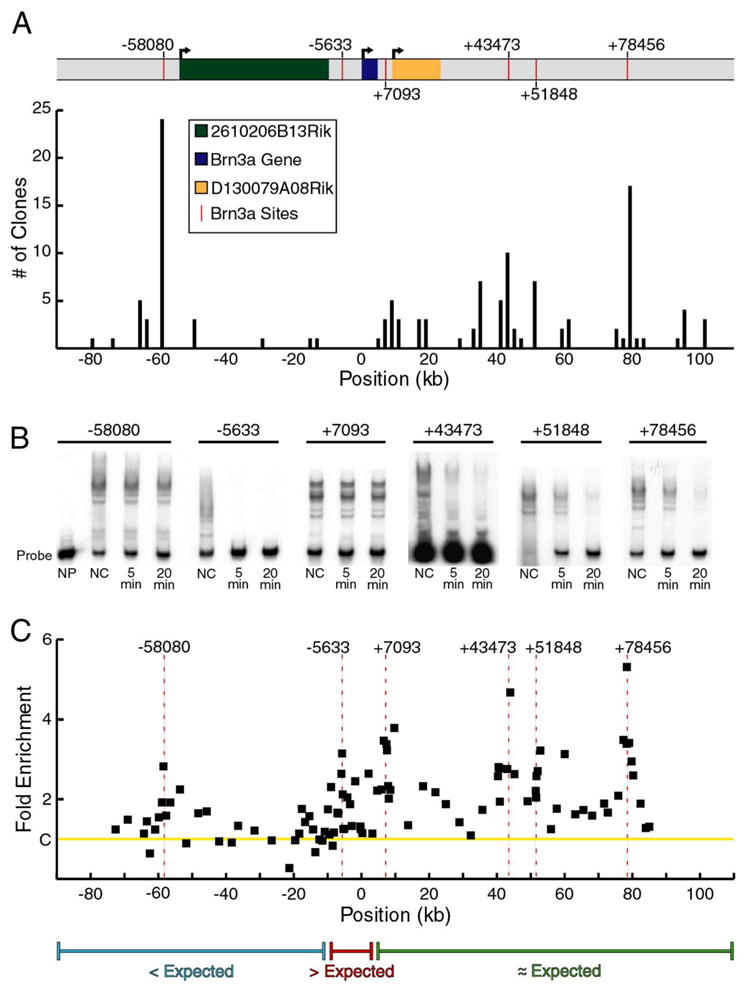
(A) Complex stability screening of a 188 kb BAC encompassing the Pou4f1 locus. BAC DNA was sheared by sonication and selected using recombinant Brn3a protein (Materials and Methods). Fragments were ligated into a cloning vector, and 128 independent clones were sequenced. The histogram indicates the number of fragments identified within each 2000 base pair interval of the BAC. In nearly every case, BAC regions identified by multiple clones contained Brn3a consensus binding sites, as indicated on the map of the Pou4f1 locus. In a prior study (Trieu, et al., 1999), lower stringency screening of a smaller region of the Pou4f1 locus identified Brn3a binding sites of moderate affinity in the autoregulatory enhancer region (−5,633). The higher stringency screening of a much larger genomic clone shown here did not identify these sites, but revealed several higher affinity sites in other regions of the locus.
Within the region encompassed by this BAC, two transcripts of unknown function have been identified, 2610206B13Rik and D130079A08Rik. These transcripts extend from −54,719 to −9,586 and +8,795 to +23,049 relative to the start of Brn3a transcription, respectively, and are transcribed on the same strand. D130079A08Rik was identified from ESTs cloned from mouse spinal ganglion, and it is unclear whether these sequences represent an independently transcribed gene, or alternate Pou4f1 transcripts.
(B) Kinetic EMSA analysis of Brn3a binding to identified Brn3a binding sites. NP, no Brn3a protein; NC, no competitor; 5 min, 5 minute incubation with competitor; 20 min, 20 minute incubation with competitor.
(C) ChIP analysis of the Pou4f1 locus using 103 primer pairs to screen a ~160 kb genomic region for in vivo binding of Brn3a. Multiple areas of enrichment were identified including the Brn3a autoregulatory site cluster at −5,633 (difference from controls, p=0.003) and four areas 3′ to the transcription unit at +7,093 (p=0.0007), +43,473 (p=0.007), +51, 848 (p<0.0001) and +78,456 (p=0.01). Binding at −58,080 did not reach significance (p>0.01). Data are average values from three ChIP assays using independent pools of E13.5 trigeminal ganglia. Primer pairs used in ChIP assays of the Pou4f1 locus appear in Table S6.
(D) A comparison of in vitro affinity and in vivo binding suggests that Brn3a binding sites 5′ to the transcription unit are occupied less often than expected based on affinity, sites in the vicinity of the autoregulatory enhancer are occupied more often than expected, and sites 3′ to the transcription unit are occupied approximately in proportion to affinity.
