Figure 2.
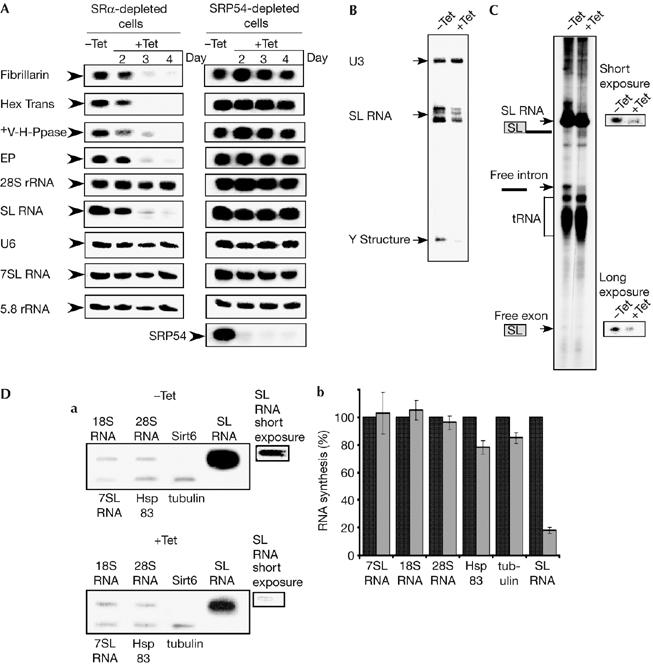
Signal-recognition particle receptor α silencing specifically inhibits spliced-leader RNA transcription and messenger RNA production. (A) The effect of SRα or SRP54 depletion on RNA level. RNA (20 μg) was prepared from uninduced (−Tet) and silenced cells 2–4 days after induction (+Tet) and subjected to northern analysis with random-labelled probes, as specified in the supplementary information online. (B) SL RNA level in SRα-depleted cells. RNA was prepared from uninduced cells (−Tet) and silenced cells 3 days after induction (+Tet) and subjected to primer extension with radiolabelled oligonucleotides complementary to the SL RNA and U3 snoRNA. Complementary DNA was separated on 6% (w/v) denaturing gel. U3, SL RNA and Y structure are indicated. (C) RNA synthesis in SRα-depleted cells. Permeable cells were prepared from the same number of cells carrying the SRα silencing construct, without induction (−Tet) or after tetracycline induction for 3 days (+Tet), as described in the Methods. The RNA was fractioned on 6% (w/v) denaturing gel. The identity of the RNAs are as indicated. (D) Slot-blot analysis of transcripts synthesized in permeable cells. (a) RNA was prepared from permeable cells, as described in (C), and was used for hybridizing with a blot carrying DNA encoding for the genes, as indicated (18S and 28S rRNA, 7SL RNA, Sirt6, tubulin, Hsp83 and SL RNA). Short and long exposures of the SL RNA signals are shown. (b) Quantitative analysis shows the percentage reduction in the level of RNA synthesis, as determined by densitometry of three independent experiments. s.d. is indicated by error bars. Black and grey bars represent uninduced and induced cells, respectively. SL RNA, spliced-leader RNA; snoRNA, small nucleolar RNA; SRα, SRP receptor α; SRP, signal-recognition particle.
