Figure 4.
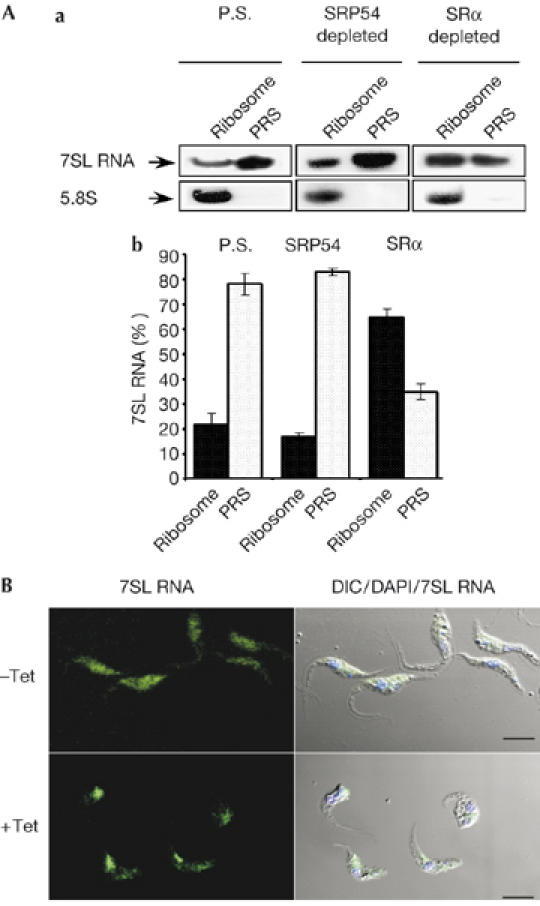
Changes in signal-recognition particle localization and ribosome association during signal-recognition parcticle receptor α depletion. (A) Binding of SRP to the RNC. (a) Extracts, PRS and ribosome fractions were prepared as described in the Methods from parental strain cells (P.S), and from cells depleted of SRP54 or SRα, 3 days after induction. RNA prepared from PRS and ribosomal fractions was subjected to northern analysis with 7SL RNA and 5.8S rRNA probes (indicated with arrows). (b) Quantitative analysis shows the percentage of 7SL RNA in PRS and ribosomal fractions, as determined by densitometry of three independent experiments. Standard deviation is indicated by error bars. (B) Localization of 7SL RNA during SRα silencing. Cells uninduced (−Tet) and after 3 days of induction (+Tet) were fixed and hybridized with DIG-labelled PCR probe to 7SL RNA, which was detected by FITC-conjugated DIG antibodies. The nucleus was stained with DAPI. Scale bars, 5 μm. DIC, differential interference contrast; DIG, digoxigenin 11; FITC, flourescein isothiocyanate; PRS, post-ribosomal supernatant; RNC, ribosome nascent chain; SRα, SRP receptor α; SRP, signal-recognition particle.
