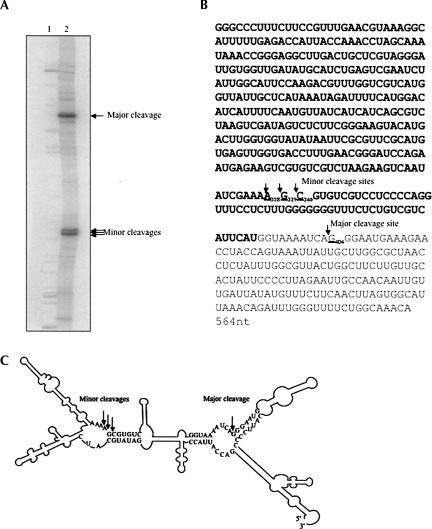
FIGURE 4.
Cleavage sites of HRA1 RNA with yeast RNase P. (A) 5′-end-labeled HRA1 RNA was a substrate for yeast RNase P cleavage in vitro. An alkali ladder and partial RNase T1 digestion of HRA1 RNA, as described in Materials and Methods, were used for comparison with the cleavage products. Yeast HRA1 RNA cleaved by yeast RNase P holoenzyme, alkali ladder of HRA1 RNA, and partial RNase T1 digestion of HRA1 RNA were individually precipitated with ethanol with glycogen as carrier, resuspended with 8 M urea dye, and loaded on to 6% polyacrylamide/8 M urea sequencing gel. After separating, the gel was dried and exposed to BioMax film (Kodak). The major and minor cleavage sites were marked with arrows. (B) The major cleavage site was located at the 5′ side of G404 (underlined), and the minor cleavage is a mixture of several cleavages at the 5′ termini of three adjacent nucleotides from A338 to G340 (bold and underlined), which were located in the antisense region that overlapped with the DRS2 gene (bold). (C) Scheme of cleavage of HRA1 RNA with major and minor cleavage sites (indicated by arrows). The secondary structure of HRA1 RNA was estimated using the MFOLD program (Zuker et al. 1999). Only individual nucleotides near the cleavage sites are shown.