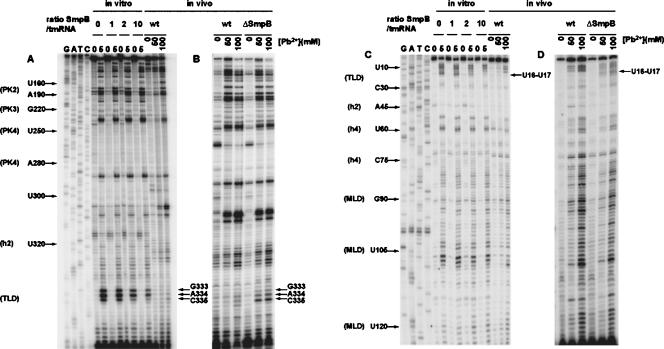
FIGURE 1.
Footprinting the tmRNA interaction with SmpB in vitro and in vivo. Lead(II) concentration for optimal (partial) cleavage in vitro was 5 mM and the SmpB:tmRNA ratios were 1:1, 2:1, and 10:1. Primer extensions with tmRNA from the wild-type (wt) strain were run on the same gels for comparison (A,C). In a parallel experiment, the same primer extensions after lead(II) cleavage were performed on total RNA extracted from isogenic wt and ΔsmpB strains (B,D). Lead(II) concentrations used for in vivo cleavage are indicated. Two different radiolabeled primers were used for primer extension analysis: GW-TM5 (A,B) and GW-TM3 (C,D). Nucleotides strongly protected from lead(II) cleavage in the presence of SmpB are indicated. G, A, T, C sequencing ladders were used for identification of cleavage positions. Some of these positions are marked to simplify their identification.