Figure 3.
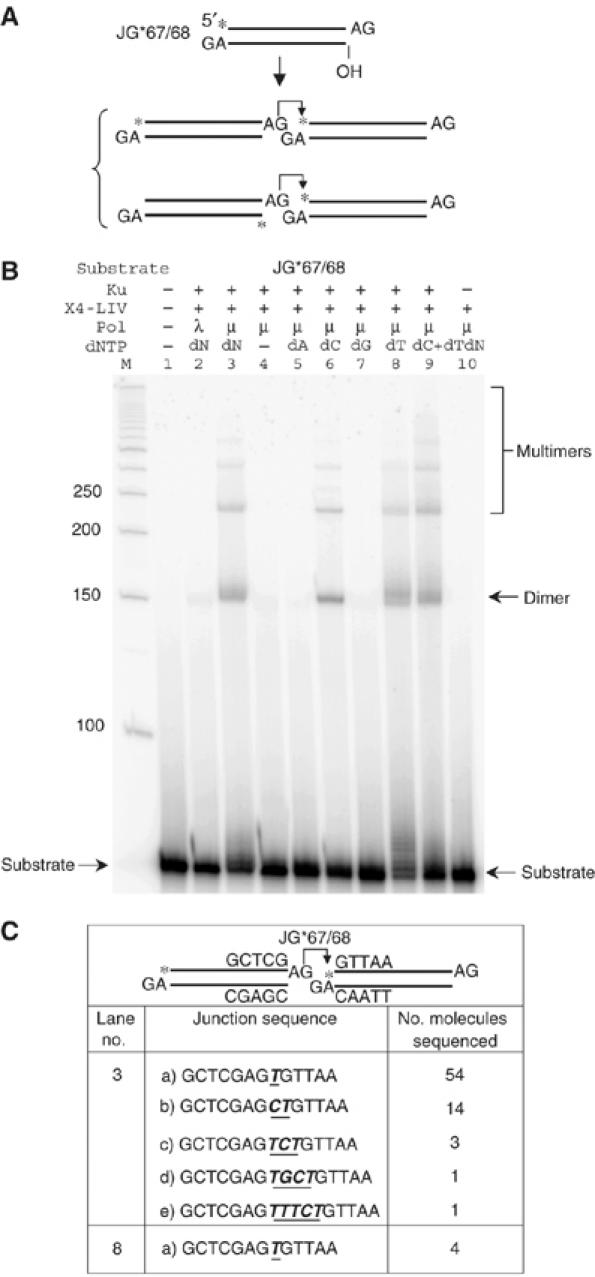
Pol mu template-independent polymerase activity provides terminal microhomology for ligation by XRCC4:DNA ligase IV. (A) The same substrate as in Figure 1A (left side) was tested for ligation. Two alternative joining pathways are proposed below the substrate. From the ligation patterns in (B) and Supplementary Figure 5, we know that the first pathway is favored. An asterisk indicates the position of the radioisotope label. (B) In each reaction, 20 nM substrate was incubated with the protein(s) indicated above each lane in a 10 μl reaction for 30 min at 37°C. After incubation, reactions were deproteinized and analyzed using 8% denaturing PAGE. Protein concentrations: Ku, 25 nM; X4-LIV, 50 nM; pol mu or lambda, 25 nM. dNTP (100 μM) was added to reactions, where indicated. ‘dN' means all the four dNTPs (100 μM each) were included. ATP (100 μM) was also added in indicated reactions. ‘M' indicates 50 bp DNA ladder. The dimer ligation product that results from the joining of two substrate molecules is labeled. Joining of more than two substrates results in trimer and higher-order species labeled as multimers. (C) Dimer products from the selected lanes were cut out of the gel, extracted, and then PCR amplified, TA-cloned, and sequenced. The junction sequences for the ligatable strand were provided. For lane 3, sequencing information was collected and combined from three individual reactions. For lane 8, two bands are apparent in the dimer product, but the longer product was not among the four molecules sequenced.
