Figure 7.
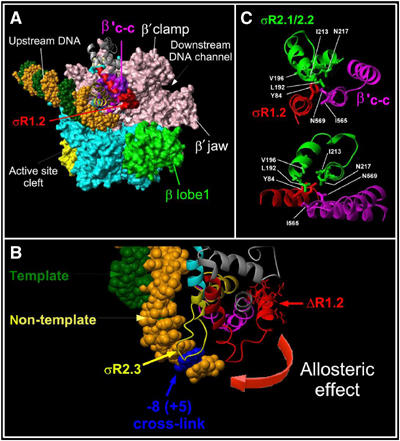
Modeling of the σ2b fragment interactions with RNAP core on the crystallographic structures of T. aquaticus and T. thermophilus RNAP. (A) The structure of T. thermophilus RNAP (PDB accession number 1IW7) fitted with the T. aquaticus RNAP structure in the complex with fork-junction DNA (PDB accession number 1L9Z). RNAP is shown as a molecular surface and DNA is shown as a CPK structure with the non-template strand in orange and the template strand in green. The large non-conserved domain present in Thermus β′ is omitted for clarity. The σ2b fragment is shown as ribbons with region 1.2 amino acids 94–101 shown as a red molecular surface. RNAP core subunits are colored as follows: β′ in rose and β in cyan; the β′ coiled-coil in violet. The β lobe that interacts with σ region 1.1 and forms the upper jaw is shown in green. (B) Modeling of σ crosslink in TEC16 on the T. aquaticus RNAP and fork-junction DNA complex structure. Color codes are as in panel A. The position corresponding to crosslinked +5G is shown in blue. (C) Interactions between σ region 1.2 (red) and 2.1–2.2 (green) and the β′ coiled-coil (magenta) in the T. thermophilus RNAP holoenzyme structure. Two projections are shown. The molecular modeling and figures were acquired using MOLMOL package (Koradi et al, 1996).
