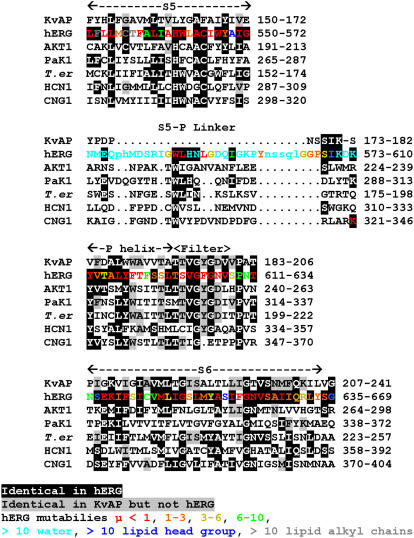
FIGURE 5.
Multisequence alignment of pore-forming domain of KvAP with hERG (ERG1, accession No. Q12809) and CNBD-containing channel homologs in plants (AKT1, accession No. NP_180233), in paramecium (PaK1, No. U19907), in Trichodesmium erythraeum bacteria ((T. er) No. YP_724331), cation nonselective HCN (No. NP_034538), and CNG (No. NP_776703) channels. The following regions are labeled on top: S5, S5-P linker, P helix, (selectivity) filter, and S6. Black background indicates residues that are identical to those of hERG, and gray background indicates residues identical to those of KvAP but not hERG. Residues of hERG are color coded according to their mutability (35) calculated from an alignment of more than 100 members of the EAG family: red μ < 1, orange μ = 1–3, yellow μ = 3–6, green μ = 6–10, cyan μ > 10 for residues predicted to be aqueous (water) exposed, blue μ > 10 for residues that occur frequently in lipid headgroup region (K, R, W, Y), gray μ > 10 for hydrophobic and ambivalent residues predicted to be exposed to lipid alkyl chains. Lower case letters in hERG S5-P linker sequence indicate positions deleted in some members of the EAG family.