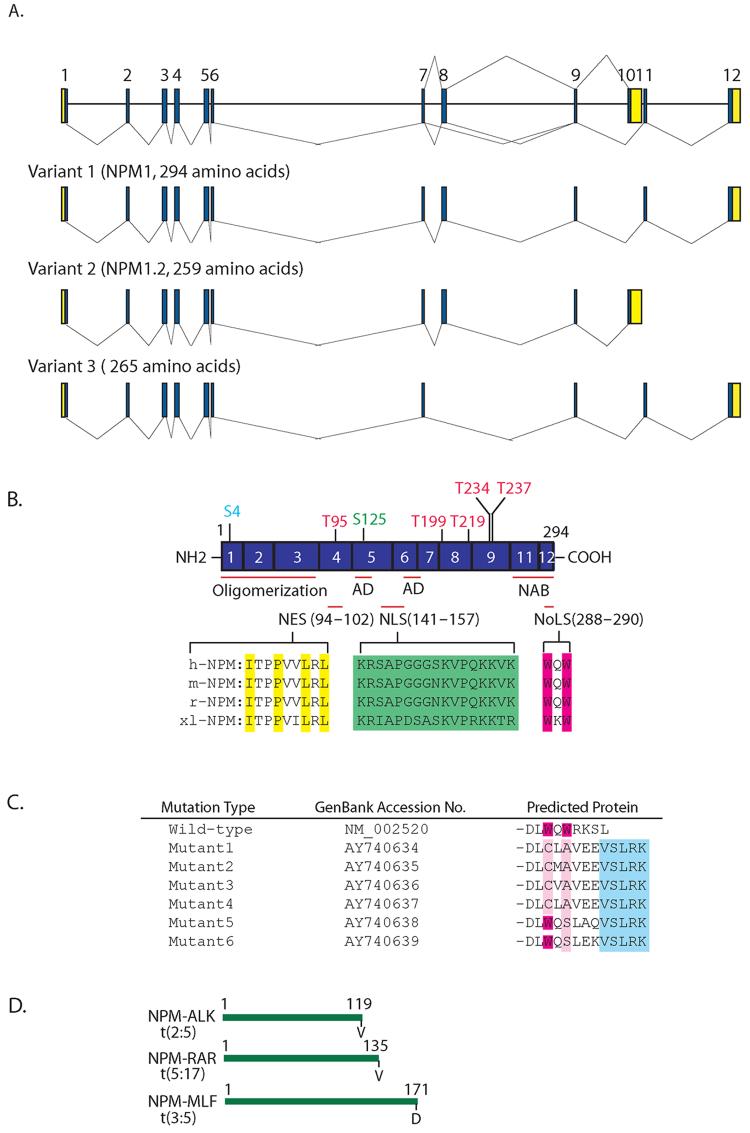
Fig. 1.
A schematic representation of the human NPM gene. (A) NPM exists as at least three isoforms, variants 1, 2 and 3. The relative sizes and positions of exons are represented by the numbers 1 .12. The line (-) represents multiple alternative splicing events. Blue and yellow areas indicate coding sequences and untranslated regions, respectively. (B) Structure of the wild-type human NPM gene. The regular-spliced NPM gene has 11 exons encoding 294 amino acids. The N-terminal and C-terminal portions of NPM are essential for oligomer formation and nucleic acid binding activity, respectively. The central region of NPM is highly acidic. AD denotes the acidic domain and NAB, the nucleic acid domain. NPM that contains several known and potential phosphorylation sites by cdc2, CKII or N-II kinase, and plk1 are shown in red, green and light blue, respectively. The NES (yellow), NLS (green), and NoLS (red) motifs are highlighted and are conserved among four different species. Prefixes: h, human; m, mouse; r, rat; xl, Xenopus laevis. (C) The C-terminus of NPM is frequently mutated in AML. The positions of the two C-terminal tryptophan (W) residues are represented by red and the mutated residues are shown in pink. The added amino acid sequences common to all the mutated proteins are represented by the light blue box. (D) The NPM N-terminal portions are translocated to ALK, RAR or MLF in lymphoma and leukemia, and their sites are also indicated.