Figure 4.
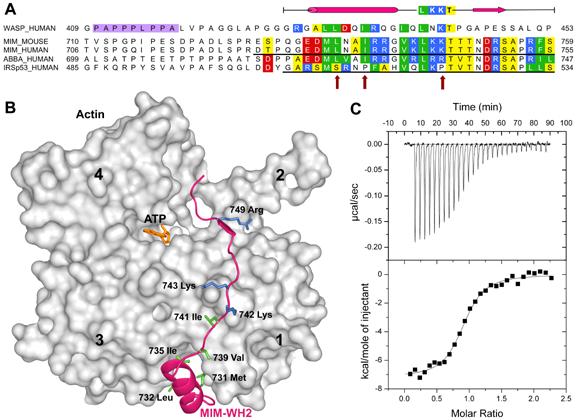
The WASP-homology domain 2 (WH2) of MIM and IRSp53. (A), Comparison of a classical WH2 (represented by WASP, Wiskott-Aldrich Syndrome Protein) with the WH2s of MIM, ABBA and IRSp53. Red, blue, green and yellow correspond to negatively charged, positively charged, hydrophobic and small (Thr, Val, Ser, Ala) conserved amino acids, respectively. The diagram above the sequences represents a secondary structure assignment based on the structure determined here (cylinder, α-helix; arrow, β-strand). Accession numbers are as in figure 1, and WASP_HUMAN, P42768. Red arrows point to non-canonical amino acids present in the WH2 of IRSp53. (B) Structure of the WH2 of MIM (red ribbon) bound to actin (gray surface). Numbers 1-4 indicate actin’s four subdomains. The side chains of some of the amino acids involved in interactions with actin are shown (green, hydrophobic; blue, positively charged). (C) Binding of the WH2 of IRSp53 to actin measured by ITC. The upper graph corresponds to the heat evolved upon repeated 10 μL injections of a 100 μM solution of the WH2 peptide into a 10 μM solution of actin in G-buffer. The lower graph shows the binding isotherm produced by integration of the heat for each injection. The line represents a nonlinear least squares fit to the data using a single-site binding model. The following thermodynamic parameters were determined from the fitting: dissociation constant Kd =0.28 ± 0.04 μM; molar enthalpy ΔH = -7.2 ± 0.1 kcal.mol-1; and stoichiometry n = 0.9.
