Abstract
The integrase from the Streptomyces phage φC31 carries out efficient recombination between the attP site in the phage genome and the attB site in the host bacterial chromosome. In this paper, we show that the enzyme also functions in human cells. A plasmid assay system was constructed that measured intramolecular integration of attP into attB. This assay was used to demonstrate that in the presence of the φC31 integrase, precise unidirectional integration occurs with an efficiency of 100% in Escherichia coli and >50% in human cells. This assay system was also used to define the minimal sizes of attB and attP at 34 bp and 39 bp, respectively. Furthermore, precise and efficient intermolecular integration of an incoming plasmid bearing attP into an established Epstein–Barr virus plasmid bearing attB was documented in human cells. This work is a demonstration of efficient, site-specific, unidirectional integration in mammalian cells. These observations form the basis for site-specific integration strategies potentially useful in a broad range of genetic engineering applications.
Genetic engineering strategies often require permanent modification of the target genome. Great sophistication typically is applied in the design of the introduced genetic material. However, cruder methods prevail for placement of the introduced gene into the genome, random integration often being state of the art. Lack of control over the position of introduced DNA results in unpredictable gene expression and potentially undesirable mutagenesis of important genes. A better solution would be a method that produces efficient site-specific integration into safe locations in the target genome.
Homologous recombination can provide great specificity in integration sites, but it occurs at too low a frequency to be optimal for genetic engineering in multicellular organisms (1). Enzymes of the site-specific recombinase family also share high specificity, and, in addition, they act with greater efficiency. Some recombinases function with no requirement for cofactors, permitting their activity in foreign cellular environments. These enzymes, isolated from a variety of microorganisms, may be ideal tools for engineering complex genomes (2). For example, the Cre recombinase from the Escherichia coli phage P1 acts efficiently in yeast, mammalian, and plant cells (3). Recombinases such as Cre, FLP, and β-recombinase (3–5) perform both integration and excision with the same target sites. Therefore, although these recombinases efficiently perform excision in mammalian cells, the net integration frequency that they mediate is low (≈0.03% for Cre; ref. 6) because of the excisive back reaction. Perhaps more ideal when integration alone is the goal are recombinases that perform only the integration reaction and require accessory factors for the reverse reaction. In this case, once integrated, the transgene cannot be excised by the recombinase. Some phage integrases have this property, for example, the integrase from the Streptomyces phage φC31 (7). Unlike the better-known λ integrase, the φC31 integrase belongs to the resolvase family of recombinases (7).
Phage integrases carry out recombination between attachment sites on the phage and bacterial genomes, known as attP and attB, respectively. For phage φC31, the attP site previously was localized to an 84-bp fragment, whereas the attB site was present on a 0.5-kb fragment (7–9). The minimal sizes of the attP and attB sites are determined in this paper. If the φC31 integrase functioned, for example, in mammalian cells and an attB site were present in the genome, it could serve as an efficient target for gene addition. The two additional requirements for integration would simply be presence of an appropriate attP site on the incoming DNA and transient presence of the integrase enzyme, which can be arranged by cointroduction of an integrase expression cassette or the protein. We demonstrate here that the φC31 integrase does indeed function in mammalian cells by using compact recognition sites. As expected, a much higher integration frequency is produced by this unidirectional integrase than by recombinases such as Cre that carry out reversible reactions.
Materials and Methods
Integrase-Expressing Plasmids.
Integrase-expressing plasmids were constructed as follows. The φC31 integrase gene was amplified by the PCR from the plasmid pIJ8600 containing the φC31 integrase and attP (a gift from Mervyn Bibb, John Innes Institute, Norwich, U.K.) with the following primers: 5′-GAACTAGTCGTAGGGTCGCCGACATGACAC-3′ and 5′-GTGGATCCGGGTGTCTCGCTACGCCGCTAC-3′. The PCR product was ligated into pCR2.1 (Invitrogen) to make the plasmid pTA-Int. The lacZ gene was removed from pCMVSPORTβGal (Life Technologies, Gaithersburg, MD) by digestion with the restriction enzymes BamHI and SpeI, and was replaced by the integrase gene from pTA-Int with BamHI- and SpeI-compatible ends, creating the plasmid pCMVInt (Fig. 1B), which expresses φC31 integrase in mammalian cells under control of the cytomegalovirus (CMV) immediate-early promoter.
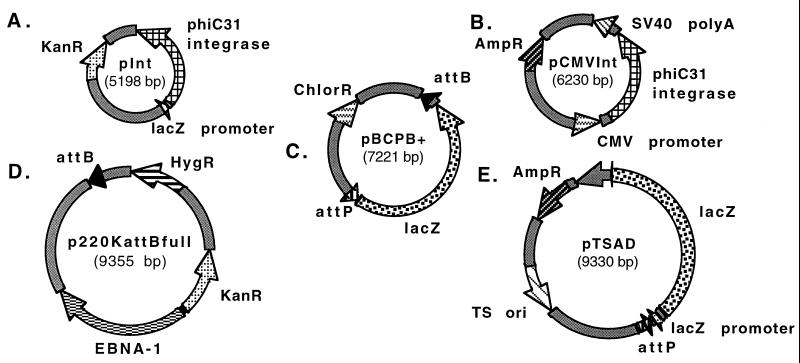
Figure 1.
Schematic diagrams of five plasmids used in demonstrating function of the φC31 integrase. (A) pInt, a plasmid for expression of φC31 integrase in E. coli. (B) pCMVInt, for expression of integrase in mammalian cells. (C) pBCPB+, intramolecular integration assay vector. (D) p220KattBfull, an EBV vector bearing attB, the target for integration events. (E) pTSAD, the donor for integration events, bears attP. KanR, AmpR, ChlorR, HygR, genes for resistance to kanamycin, ampicillin, chloramphenicol, and hygromycin, respectively; SV40, simian virus 40; EBNA-1, Epstein–Barr virus-encoded nuclear antigen 1.
A BamHI/PstI fragment from pCMVInt containing the integrase gene was cloned into pACYC 177 (with resistance to ampicillin and kanamycin; a gift of Stanley N. Cohen, Stanford; ref. 10) treated with BamHI and PstI to remove part of the ampicillin-resistance gene. Finally, the lacZ promoter was removed from pBCSK+ (Stratagene) by digestion with SacI and SapI. The integrase-containing pACYC plasmid was digested with PstI and SacI, and the lacZ promoter was inserted upstream of the integrase gene with a linker (5′-GCTCGGCCAAAAAGGCCTGCA-3′ and 5′-GGCCTTTTTGGCCG-3′), creating the plasmid pInt (Fig. 1A), expressing the φC31 integrase under control of the lacZ promoter.
Intramolecular Integration Assay in E. coli.
The intramolecular integration assay plasmid was constructed as follows. A 285-bp fragment containing the bacterial attachment site for φC31 (attB) was amplified by PCR from Streptomyces lividans genomic DNA (a gift of Stanley N. Cohen) with the primers 5′-CAGGTACCGTCGACGATGTAGGTCACGGTC-3′ and 5′-GTCGACATGCCCGCCGTGACCG-3′. This fragment was ligated into pCR2.1 to create the plasmid pTA-attB. The phage attachment site (attP) was amplified by PCR from pIJ8600 with the primers 5′-CGACTAGTACTGACGGACACACCGAA-3′ and 5′-GTACTAGTCGCGCTCGCGCGACTGACG-3′ and was ligated into pCR2.1 to create the plasmid pTA-attP, containing a 221-bp attP region. The lacZα region was removed from pBCSK+ by digestion with PvuI and KpnI, treatment with T4 polymerase, and religation. The full-length lacZ gene from pCMVSPORTβGal was removed by digestion with SpeI and HindIII and cloned into the SpeI and HindIII sites of the lacZα-deficient pBCSK+ to make pBCβGal. The attP was then removed from pTA-attP by SpeI digestion and cloned into the SpeI site of pBCβGal to create pBCβGal-attP. The attB was removed from pTA-attB by SalI digestion and cloned into the SalI site of the attP-containing pBCβGal-attP to create the assay plasmid pBCPB+ (Fig. 1C), in which the TTG cores of the two att sites are in the same orientation. In addition, a control plasmid, pBCPB−, in which the att sites are in opposite orientations, was also constructed.
The pInt plasmid was then transformed into DH10B bacteria. Cells were grown under kanamycin selection and made electrocompetent by a standard protocol. The resulting electrocompetent DHInt cells were used in the bacterial intramolecular integration assay, which was conducted as follows. A 0.2-ng sample of the assay plasmid of choice was electroporated into DHInt cells, which were allowed to recover for 1 h, spread on LB plates containing 25 μg/ml chloramphenicol, 60 μg/ml kanamycin, and 50 μg/ml 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal), and grown at 37°C. The lac inducer isopropyl β-d-thiogalactoside (IPTG) was not present or necessary. If an intramolecular integration event occurs, the lacZ gene located between the attB and attP sites will be excised, and a resulting colony will be white. The frequency of intramolecular integration was calculated as the number of white colonies divided by the total number of colonies × 100.
Intramolecular Integration Assay in Human Cells.
Subconfluent (60–80%) 60-mm plates of human 293 cells (American Type Culture Collection; ref. 11) grown in DMEM supplemented with 9% FBS and 1% penicillin/streptomycin were transfected with Lipofectamine (Life Technologies) at a ratio of 6 μg of Lipofectamine per μg of DNA. Experiments were performed with 100 ng of the assay plasmid of interest and 2 μg of pCMVInt. Negative controls performed in each experiment included no DNA, pCMVInt only, pBCβgal (assay plasmid with no att sites), pBCβgal + pCMVInt, and pBCPB+ alone.
At 24 h after transfection, the medium was supplemented with 50 units/ml DNase I to reduce the background of untransfected DNA. Low molecular weight DNA was recovered 72 h after transfection as described by Hirt (12). A portion of this DNA was electroporated into competent DH10B E. coli cells and spread on plates containing chloramphenicol and X-Gal to select for the assay plasmid only. The intramolecular integration frequency was determined to be the number of white colonies divided by the total number of colonies × 100.
PCRs on Hirt extracts contained 400 nM forward (5′-GGCGAGAAAGGAAGGGAAGA-3′) and T3 (5′-ATTAACCCTCACTAAAGGGA-3′) primers, 1.5 units of Taq DNA polymerase, 10 mM Tris⋅HCl (pH 9), 50 mM KCl, 1.5 mM MgCl2, 200 μM dNTPs, and either 250 pg of plasmid DNA as a control or 1/40 fraction of a Hirt extract from a 60-mm plate. Reactions were overlaid with mineral oil, and PCR was conducted as follows: 94°C for 5 min, followed by 35 cycles of 94°C for 30 sec, 62°C for 30 sec, and 72°C for 45 sec. A final extension period of 72°C for 7 min then was performed. PCR products were analyzed on 2% agarose (NuSieve 3:1; FMC) in TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3).
Unreacted pBCPB+ plasmid DNA was used as a negative control in the PCR. The positive recombination control was generated from pBCPB+ in vitro by incubating plasmid DNA with a crude protein extract from integrase-expressing E. coli. In vitro recombination reactions contained 100 ng of pBCPB+, 1 μg of integrase extract, 20 mM Tris⋅HCl (pH 7.5), 100 mM NaCl, 0.1 mM EDTA, and 1% glycerol in a final volume of 20 μl. Reactions were assembled on ice, incubated at 37°C for 1 h, and heat-inactivated at 80°C for 20 min. Crude extract was obtained from BL21-SI E. coli-expressing integrase (Life Technologies) cloned into vector pET11 (Novagen).
Plasmids for Intermolecular Integration Assay.
The attB and attP plasmids needed for this study were constructed as follows. The target hygromycin-resistant Epstein–Barr virus (EBV)-based plasmids were based on p220.2 (13). The negative control plasmid p220Kan was made by inserting the kanamycin-resistance gene from the Kan-resistant Genblock (Amersham Pharmacia) into the XmnI site of the ampicillin-resistance gene of p220.2. To make attB-containing p220 plasmids, the ampicillin-resistance gene of p220.2 was removed by digestion with BspHI. The kanamycin-resistance gene described above was isolated by digestion with PstI and cloned into amp-deleted p220.2 with BspHI/PstI linkers (5′-CATGAGGCCAAAAAGGCCTGCA-3′ and 5′-GGCCTTTTTGGCCT-3′) to create the plasmid p220K. The full-length attB was removed from the plasmid pTA-attB by SalI digestion and cloned into the SalI site of p220K, creating the plasmid p220KattBfull (Fig. 1D). The 35-bp attB was cloned into the SalI and BamHI sites of p220K by using the oligonucleotides 5′-GATCCGATATCGCGCCCGGGGAGCCCAAGGGCACGCCCTGGCACCG-3′ and 5′-TCGACGGTGCCAGGGCGTGCCCTTGGGCTCCCCGGGCGCGATATCG-3′, creating the plasmid p220KattB35.
These EBV plasmids, p220Kan, p220KattBfull, and p220KattB35, were established in human 293 cells as follows. Human 293 cells were grown in DMEM containing 9% FBS and 1% penicillin/streptomycin to ≈70% confluency in 100-mm plates. Samples (8 μg) of p220KattBfull, p220Kattb35, or the control p220Kan were introduced by transfection with Lipofectamine according to the manufacturer's protocol. At 24 h posttransfection, the cells were split 1:4, and at 48 h posttransfection, hygromycin selection (350 μg/ml) was begun. At 11 to 14 days after starting selection, the cells were expanded and frozen.
The attP-containing plasmid pTSAD (Fig. 1E) was constructed as follows. A multiple cloning site (oligos 5′-AATTACCGCGGGGCGCGCCGTTTAAACGCATGCCAATTGGGCCGGCCG-3′ and 5′-AATTCGGCCGGCCCAATTGGCATGCGTTTAAACGGCGCGCCCCGCGGT-3′) was cloned into the EcoRI site of the plasmid pWTLox2 (18) upstream of lacZ, regenerating one EcoRI site. The attP site was removed from the plasmid pTA-attP by digestion with EcoRI and cloned into the regenerated EcoRI site of pWTLox2 to create the plasmid pES1. The lacZ promoter was removed from pBCSK+ by digestion with PvuII and SacII and cloned into pES1 that had been digested with PmeI and SacII. The region containing attP, the lacZ promoter, and the lacZ gene was removed by digestion with BamHI and BglII and cloned into the BamHI site of pTSA30 (a gift of Gregory Phillips, Iowa State University, Ames; ref. 14) to create the donor plasmid pTSAD. pTSA30 and its pTSAD derivative are temperature-sensitive for plasmid replication in E. coli.
Intermolecular Integration Assay in Human Cells.
To perform the integration assay, EBV plasmid-containing cells were grown to confluency in DMEM containing 9% FBS, 1% penicillin/streptomycin, and 200 μg/ml hygromycin in 100-mm plates. These plates were split into eight 60-mm plates and grown in the above medium without hygromycin for 24–48 h, until they were ≈60–80% confluent. pCMVInt (Fig. 1B) and pTSAD were transfected in equimolar amounts (10 μg of total DNA) by using 50 μl of Superfect (Qiagen, Valencia, CA), according to the manufacturer's protocol. As negative controls, no DNA, 4 μg of pCMVInt, or 6 μg of pTSAD was cotransfected with salmon sperm DNA to 10 μg. In addition, an equimolar amount of a plasmid encoding the green fluorescent protein gene under control of the CMV promoter (a derivative of pEGFP-C1; CLONTECH) with salmon sperm DNA to 10 μg was transfected in parallel into the EBV plasmid-containing cells to monitor transfection efficiency. Transfected cells were trypsinized, suspended in PBS, and placed on slides, and several fields were counted by eye under a fluorescence microscope.
Cells were grown in medium containing serum and 50 units/ml DNase I at 24 h after transfection, and they were harvested 72 h after transfection. Low molecular weight DNA was purified by Hirt extraction and transformed into DH10B E. coli by electroporation. Also, 24 h after transfection, transfection efficiency was measured by counting the green fluorescent protein-expressing cells relative to the total number of cells. The transfection efficiencies typically ranged from 6% to 18%. Because untransfected cells would have no opportunity to undergo integration but would still contribute EBV plasmids to the bacterial assay in the form of white colonies, the transfection efficiency was used to obtain the correct integration frequency.
In a typical experiment, 15 μl of a transformation was spread on each of three plates containing 60 μg/ml kanamycin, 50 μg/ml X-Gal, and 10 μg/ml isopropyl β-d-thiogalactoside (IPTG), and 150 μl of the same transformation was spread on each of three plates containing 100 μg/ml ampicillin, 100 μg/ml methicillin, 60 μg/ml kanamycin, 50 μg/ml X-Gal, and 10 μg/ml IPTG. The bacteria were grown overnight at 42°C for ≈16 h. The elevated temperature prevented replication of pTSAD, which has a temperature-sensitive plasmid origin of replication. Integrants were scored as blue colonies on the plates containing both kanamycin and ampicillin. Integration frequency was calculated as the number of blue colonies on kanamycin and ampicillin plates divided by (the total number of colonies on kanamycin plates × 10) for each set of transfections. The colony number on kanamycin plates was multiplied by 10 because only 10% of the transformation was spread on these plates, relative to the amount spread on kanamycin and ampicillin plates. Raw numbers for integration frequency were divided by transfection efficiency to obtain accurate values for integration frequency.
Results
Intramolecular Integration Assay in E. coli.
To create a simple and rapid assay for integration, we developed a plasmid that would provide a quantitative measure of intramolecular integration frequency. The pBCPB+ plasmid carries the φC31 attP and attB sites in direct orientation flanking a lacZ gene on a chloramphenicol-resistant ColE1 derivative (Fig. 1C). The plasmid carries a 221-bp region derived from φC31 DNA and known to contain the attP site (9). Because an 84-bp subregion from within this fragment gives good integration function (7), it was likely that the 221-bp fragment contained the complete attP site. Plasmid pBCPB+ also contains a region of ≈285 bp known to contain the attB site, obtained by PCR from the S. lividans genome. The φC31 integrase gene, under the control of the lacZ promoter, was placed on a kanamycin-resistant derivative of pACYC177 to form pInt (Fig. 1A). These two plasmids, pBCPB+ and pInt, belong to different compatibility groups and can therefore coexist in E. coli.
When the pBCPB+ assay plasmid is transformed into an E. coli strain, DHInt, in which the pInt plasmid is resident, phage integrase will catalyze integration of attP into the attB site, resulting in excision of the lacZ gene. This integration event consequently produces a color change from blue to white when the bacteria form colonies on agar plates containing X-Gal.
When this assay was carried out in DHInt bacteria carrying pInt by using pBCPB+, all colonies were white, indicating efficient integration (Table 1). The same plasmid produced only blue colonies in DH10B bacteria, in the absence of the integrase gene. These results verify that our assay plasmid carries functional attB and attP sites and that the φC31 integrase functions efficiently in E. coli with no added cofactors. In contrast, the plasmid pBCPB−, which carries the att sites in inverted orientation, resulted in blue colonies, because the lacZ gene was merely inverted, not excised, by the integration reaction. The assay plasmid with no att sites, pBCβgal, also yielded only blue colonies in DHInt cells. Restriction-enzyme digestion of plasmid DNA purified from a representative number of white colonies verified that the intramolecular integration reaction occurred as expected and resulted in the deletion of lacZ between the attB and attP sites. DNA sequencing of an integrant demonstrated the expected exact recombination reaction mediated by the integrase.
Table 1.
Intramolecular integration frequencies
| Cells | Plasmid | n | Colonies
|
Intramolecular integration frequency, % | |
|---|---|---|---|---|---|
| White | Total | ||||
| DHInt E. coli | None | 0 | 0 | 0 | NA |
| pBCβgal | 3 | 0 | 2,369 | <0.04 | |
| pBCPB− | 1 | 0 | 1,398 | <0.07 | |
| pBCPB+ | 3 | 2,284 | 2,284 | 100 | |
| 293 Human | None | 8 | 0 | 0 | NA |
| pInt | 8 | 0 | 0 | NA | |
| pBCβgal | 8 | 5 | 6,090 | 0.08 | |
| pBCβgal and pInt | 8 | 30 | 7,643 | 0.39 | |
| pBCPB+ | 8 | 7 | 4,403 | 0.16 | |
| pBCPB+ and pInt | 16 | 6,971 | 13,293 | 52.44 | |
Numbers of blue and white colonies and integration frequencies for plasmids assayes in DHInt E. coli and for plasmids recombined in human 293 cells and assayed in DH10B. n, Number of independent experiments for each plasmid; NA, not applicable.
Intramolecular Integration Assay in Human Cells.
To test whether the φC31 integrase can function in a mammalian cell environment, we transfected pBCPB+ and an integrase expression plasmid, pCMVInt (Fig. 1B), that carries the CMV promoter active in mammalian cells, into human 293 cells. After 72 h, plasmid DNA was purified from the cells by Hirt extraction (12) and transformed into bacteria for scoring. Using this assay system in mammalian cells, we determined that the φC31 integrase catalyzed recombination between the full-length attB and attP sites of pBCPB+ at a frequency of 52.4% (Table 1; mean of 16 experiments, SE = 2.32%). This frequency is likely to be an underestimate, because plasmid DNA that never came in contact with the φC31 integrase was probably present, despite our efforts to remove untransfected DNA with DNase I.
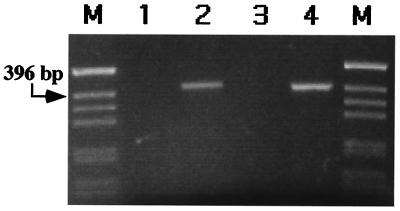
To assure that this recombination was happening in the human cells and not later in the bacteria, pCMVInt and pBCPB+ were coelectroporated into DH10B E. coli. In this control, 1.08% white colonies were observed, verifying that the vast majority of the recombinants occurred in the human cells. To further verify that the site-specific recombination was occurring in the mammalian cells, low molecular weight DNA isolated by Hirt extraction from the human cells was subjected to PCR with specific primers that would amplify a 401-bp product only from the recombinant. These experiments revealed the expected fragment only when both pBCPB+ and pCMVInt were cotransfected into the human cells (Fig. 2). We determined the DNA sequence of this PCR fragment, revealing that the recombination occurred as expected for integrase-mediated site-specific recombination between attP and attB.
Figure 2.
PCR analysis of recombination in human cells. A 401-bp product is indicative of site-specific recombination. Lane 1, untransfected pBCPB+; lane 2, in vitro-reacted pBCPB+ (see Materials and Methods); lane 3, Hirt extract (12) containing transfected pBCPB+ from human 293 cells; lane 4, Hirt extract containing transfected pBCPB+ and pCMVInt from human 293 cells; lanes M, size markers.
To verify site-specific recombination, 96 of the white colonies were picked, and plasmid DNA was prepared and examined by restriction digestion. Of these, 97% contained a plasmid that represented the expected site-specific recombinant. The remaining colonies contained plasmids that carried large rearrangements that disrupted lacZ. The low-frequency rearrangement of transfected plasmids has been observed with all plasmids, with and without integrase and att sites, and can be attributed to transfection-associated mutation of newly introduced DNA (15). This result indicates that the φC31 integrase is active in mammalian cells and efficiently carries out the integration reaction when transiently introduced with its att sites.
Determination of Minimal Sizes of attB and attP.
Before this study, the minimal sizes for the φC31 attachment sites attB and attP had not been reported. The attB site had been localized to ≈280 bp, and the attP region had been localized to 84 bp (7). We used the intramolecular integration assay described above to determine the minimal functional sizes for these att sites. Short double-stranded adaptor molecules containing att sites of various lengths were created by annealing single-stranded oligonucleotides. These shorter sites were used to replace the full-length att sites in the pBCPB+ assay plasmid, and recombination efficiencies were determined by electroporation into E. coli.
To determine the minimal functional size of attB, the BamHI and HindIII fragment of pBCβGal-attP was replaced by a series of synthetic shorter sites having ends permitting their orientation-appropriate cloning. The resulting plasmids were electroporated into DHInt E. coli cells, and recombinants were scored as white colonies, as described above. Fig. 3 (Upper) shows the results of these experiments. attB sites of 50, 40, 35, and 34 bp all provided full recombination function in this assay, i.e., they functioned at 100% of the efficiency of the full-length attB. Reduction of the site to 33 bp produced a marked decrease in recombination activity, so we concluded that 34 bp is the minimal functional size of attB. This determination is somewhat arbitrary, because under more stringent reaction conditions, such as less time, sites of 34 bp in length might be revealed to function less well than does the full-length site. However, 34 bp clearly provides a substantial level of function and probably retains most of the critical sequences. In addition, a site of this length encompasses the major inverted repeats present in the attB region (6).
Figure 3.
Minimal sizes of attB and attP sites. The right column shows the percentage recombination obtained in the intramolecular integration assay in E. coli when the illustrated shortened versions of φC31 attB (Upper) and attP (Lower) were tested. The name of each site tested corresponds to the length of the att site in base pairs. B33A and B33B indicate sites where the reduction of the site length from 34 bp to 33 bp occurred at the right or left ends of the site, respectively. Similar nomenclature is used for P39A and P39B. full attP, full-length attP.
Once attB was determined to be 34 bp long, attP was subjected to a similar set of reductions. The reduced attP sites were assayed in a plasmid carrying attB34 rather than full-length attB. To perform these experiments, the full-length attP surrounded by SacII and SpeI sites was replaced with a series of synthetic annealed oligonucleotides bearing ends that permitted their correct orientation-specific cloning into pBCPBattB34+. Fig. 3 (Lower) depicts the results of these experiments. The function of attP dropped off as its size was reduced from 40 to 36 bp. The DNA sequence reveals that the 38-bp site encompasses the major inverted repeat evident in attP, but it is apparent from this data that one of the next outermost base pairs conveys some function (P39B). From this analysis, we conclude that the minimal size of attP is 39 bp, subject to the same caveats discussed previously for attB.
To determine the frequency at which the reduced att sites function in mammalian cells, the same panel of plasmids was transfected into human 293 cells along with pCMVInt. After 72 h in mammalian cells, plasmid DNA was purified by the method of Hirt (12) and transformed into DH10B E. coli cells for scoring of recombinants. The results of these experiments showed that minimal sizes for attB and attP similar to those determined in E. coli also apply in mammalian cells (data not shown). We were able to achieve ≈60–90% of the efficiency of the full-length att sites with the same reduced att sequences that worked at 100% efficiency in E. coli, probably because the overall reaction is somewhat less efficient in the mammalian cell environment. The lesser frequencies perhaps reveal with more sensitivity that the minimal sites identified in the E. coli assay do lack some functional activity.
Bimolecular Integration Assay into a Model Chromosome in Mammalian Cells.
We showed that the φC31 integrase efficiently catalyzes site-specific intramolecular integration in mammalian cells. We wanted to test whether the integrase could catalyze efficient site-specific integration of exogenous DNA into mammalian chromosomes in cell culture. EBV-based plasmids are good models for chromosomes and are much easier to work with. EBV vectors are retained in the nucleus, replicate in synchrony with the chromosomes, and bear chromatin indistinguishable from that of the chromosomes (16). They can easily be purified from cells and transformed into E. coli for rapid scoring of integration events, thus they have great utility in characterization of the integration reaction in human cells.
In these experiments, one kanamycin-resistant EBV plasmid was equipped with the full attB site (Fig. 1D) and another with a minimal attB site (attB35), and each was established in human 293 cells to create a stable attB-containing human cell line. An ampicillin-resistant plasmid carrying attP and lacZ and having a temperature-sensitive origin of replication (pTSAD; Fig. 1E) was then cotransfected into the attB cell line, along with a plasmid expressing the φC31 integrase. To assay for integration products, after 3 days, plasmid DNA was extracted, transformed into bacteria, and grown at the nonpermissive temperature. Blue colonies that grew on plates containing kanamycin, ampicillin, and X-Gal were scored as integrants, whereas total colony number could be obtained by plating on kanamycin alone.
Table 2 lists the integration frequencies obtained with each EBV test plasmid and the negative controls. Each line of the table represents a minimum of three separate transfections. For p220Kan, which lacks the attB site, a negligible frequency of blue colonies was detected. On restriction analysis, these plasmids were determined not to be integrants. Some appeared to represent homologous recombination events that occurred through common amp sequences on the two plasmids. For p220KattB35, carrying a smaller attB, significant numbers of blue colonies were detected. When corrected for the transfection efficiency in these experiments, the integration frequency was 1.7%. For p220KattBfull, the integration frequency was even higher at 7.5%. This increase presumably reflects a favorable sequence context for the full attB site as compared with the reduced site. Controls in which pCMVInt, pTSAD, and each of the EBV plasmids, p220Kan, p220KattBfull, and p220KattB35, were cotransformed directly into E. coli yielded negligible numbers of blue colonies (0.002% or less). These controls confirmed that the high frequency of integration events scored above occurred in human cells, not in E. coli.
Table 2.
Bimolecular integration assay in human cells
| Plasmid | Blues/total | % Integration | Corrected % integration |
|---|---|---|---|
| p220Kan | |||
| No DNA | 0/7,970 | <0.01 | <0.08 |
| pCMVInt only | 0/13,600 | <0.007 | <0.04 |
| pTSAD only | 1/11,830 | 0.008 | 0.05 |
| pCMVInt + pTSAD | 3/32,680 | 0.009 | 0.06 |
| p220KattB35 | |||
| No DNA | 0/64,530 | <0.002 | <0.01 |
| pCMVInt only | 0/42,720 | <0.002 | <0.02 |
| pTSAD only | 0/39,930 | <0.003 | <0.02 |
| pCMVInt + pTSAD | 382/157,710 | 0.242 | 1.69 |
| p220KattBfull | |||
| No DNA | 0/70,350 | <0.001 | <0.01 |
| pCMVInt only | 0/41,960 | <0.002 | <0.02 |
| pTSAD only | 0/39,740 | <0.003 | <0.02 |
| pCMVInt + pTSAD | 1,799/166,890 | 1.08 | 7.50 |
Results are shown for human cells carrying one of three EBV plasmids, p220Kan, a negative control lacking attB; p220KattB35, which carries a similar attB; and p220KattBfull, carrying the full-sized attB. Integration frequencies are shown for experiments when no DNA was transfected, when either the integrase expression plasmid pCMVInt or the attP-bearing plasmid pTSAD were transfected alone, or when both pCMVInt and pTSAD were transfected together. Only the latter condition, in the presence of a plasmid bearing attB, led to integration events. Integration frequencies were corrected for transfection frequency to give the accurate corrected integration frequencies in the last column. p220KattBfull produced the highest integration frequency at 7.5%.
Furthermore, the integrants are site-specific, as indicated by restriction mapping of >160 of the blue colonies from the experiments with p220KattB35 and p220KattBfull. In addition, two integrants each from the experiments with p220Katt35 and p220Kattfull were analyzed at the DNA sequence level across the junctions of the integration site, confirming that exact site-specific integration occurred between attB and attP. Table 2 indicates that, as expected, the reaction requires presence of both the integrase gene (pCMVInt) and the attP site (pTSAD). Because EBV vectors are nuclear, chromatinized minichromosomes, the high integration frequency obtained in this system may be predictive of integration frequencies into att sites located on the chromosomes.
Discussion
This study demonstrates that the φC31 integrase, derived from a Streptomyces phage, can function efficiently in mammalian cells. The demonstration that the enzyme could function in E. coli and that purified φC31 integrase was active in an in vitro system implied a lack of requirement for Streptomyces-specific cofactors (7). On this basis, we hypothesized that the enzyme might have the ability to function in mammalian cells. It was necessary to express the enzyme in a mammalian cell environment and demonstrate activity in integration assays to provide evidence that the enzyme could function in higher eukaryotic cells. In this way, pairing and reacting attP and attB sites in the context of chromatin, large genome size, and other features distinct from the prokaryotic environment where φC31 integrase had previously been shown to work, were tested.
By placing the attB sequence on a stable chromatinized EBV plasmid established in human cells, then demonstrating the specific and efficient integration of a plasmid carrying an attP site into attB, we have provided proof that the enzyme can locate and recombine its recognition sites in mammalian cells. This finding suggests that the enzyme may mediate chromosomal integration in a wide range of host environments, including plants, insects, other mammals, and diverse microorganisms.
Our demonstration of 34-bp and 39-bp minimal sizes for the attB and attP sites, respectively, makes it unlikely that they will be present even in the large genomes of mammals and most plants. However, the enzyme may be able to mediate integration at naturally occurring pseudo-att sites in eukaryotic genomes having significant similarity to native att sequences (18). The expected rarity of good matches with att sites may limit recombination to a small number of chromosomal pseudo-att sites, which may produce usable integration frequencies at endogenous locations in the chromosomes.
This integration system is significantly more efficient and specific than currently available alternatives. The integration frequency into an attB site located on an EBV plasmid is several orders of magnitude higher than the frequency of random integration (1). The site-specificity of the enzyme also distinguishes it from the random integration mediated by retroviral integrases and most transposases. With regard to other recombinases, because there is no accompanying excision reaction with the φC31 integrase, the integration frequency is more than two orders of magnitude higher than that of the Cre recombinase (6). Recently, integration mediated by the integrase protein of lambdoid phage HK022 has been reported in mammalian cells (17). This integrase requires host cofactors in E. coli and seems to produce a net integration frequency in mammalian cells at least an order of magnitude lower than that of the φC31 integrase. The high integration frequency mediated by an autonomous unidirectional integrase such as that of φC31 may enable a variety of genomic modifications desirable for research, commercial, or therapeutic purposes.
Acknowledgments
We thank Drs. Mervyn Bibb, Stanley N. Cohen, and Gregory Phillips for plasmids and DNA. This work was supported by grants from the National Institutes of Health (DK51834) and the Cystic Fibrosis Foundation (to M.P.C.). E.C.O. was supported by a Ford Foundation predoctoral fellowship. A.C.G. was supported by a predoctoral fellowship from Public Health Service National Research Service Award 5 T32 CA09302 from the National Cancer Institute.
Abbreviations
- EBV
Epstein–Barr virus
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
- CMV
cytomegalovirus
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.090527097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.090527097
References
- 1.Vega M A. Hum Genet. 1991;87:245–253. doi: 10.1007/BF00200899. [DOI] [PubMed] [Google Scholar]
- 2.Stark W M, Boocock M R, Sherratt D J. Trends Genet. 1992;8:432–439. [PubMed] [Google Scholar]
- 3.Sauer B. Curr Opin Biotechnol. 1994;5:521–527. doi: 10.1016/0958-1669(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 4.Diaz V, Rojo F, Martinez-A C, Alonso J C, Bernad A. J Biol Chem. 1999;274:6634–6640. doi: 10.1074/jbc.274.10.6634. [DOI] [PubMed] [Google Scholar]
- 5.O'Gorman S, Fox D T, Wahl G M. Science. 1991;251:1351–1355. doi: 10.1126/science.1900642. [DOI] [PubMed] [Google Scholar]
- 6.Sauer B, Henderson N. New Biol. 1990;2:441–449. [PubMed] [Google Scholar]
- 7.Thorpe H M, Smith M C M. Proc Natl Acad Sci USA. 1998;95:5505–5510. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhstoss S, Rao R N. J Mol Biol. 1991;222:897–908. doi: 10.1016/0022-2836(91)90584-s. [DOI] [PubMed] [Google Scholar]
- 9.Rausch H, Lehmann M. Nucleic Acids Res. 1991;19:5187–5189. doi: 10.1093/nar/19.19.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang A C Y, Cohen S N. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham F L, Smiley J, Russell W C, Nairn R. J Gen Virol. 1977;36:59–72. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 12.Hirt B. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 13.DuBridge R B, Tang P, Hsia H C, Leong P M, Miller J H, Calos M P. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phillips G J. Plasmid. 1999;41:78–81. doi: 10.1006/plas.1998.1380. [DOI] [PubMed] [Google Scholar]
- 15.Lebkowski J S, DuBridge R B, Antell E A, Greisen K S, Calos M P. Mol Cell Biol. 1984;4:1951–1960. doi: 10.1128/mcb.4.10.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haase S B, Calos M P. Nucleic Acids Res. 1991;19:5053–5058. doi: 10.1093/nar/19.18.5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolot M, Silberstein N, Yagil E. Mol Biol Rep. 1999;26:207–213. doi: 10.1023/a:1007096701720. [DOI] [PubMed] [Google Scholar]
- 18.Thyagarajan B, Guimaraes M J, Groth A C, Calos M P. Gene. 2000;244:47–54. doi: 10.1016/s0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]