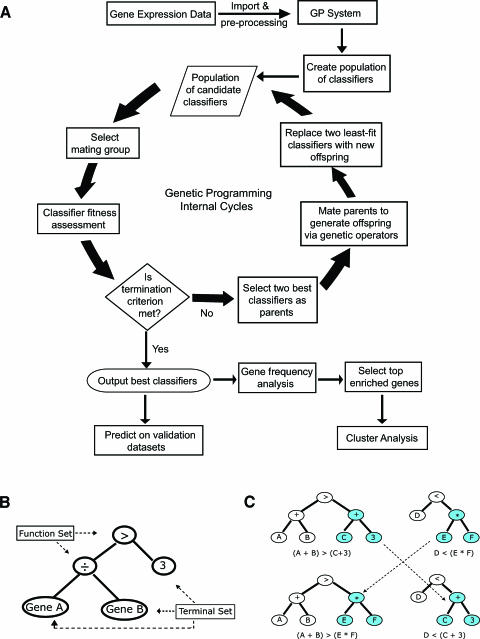
Figure 1.
(A) Flowchart for the GP process. Briefly, a population of tree-based classifiers is first created by randomly choosing gene expression data or constant values and combining with arithmetic or Boolean operators. An example of tree-based classifiers is represented in (B). A small subgroup of classifiers is then selected as a “mating group” and each classifier in this mating group is assessed by a fitness function, which is defined as the area under the ROC-AUC in this study. The two fittest classifiers are then selected as “mating” parents and “mated” to produce “offspring” by genetic operators (crossover or mutation). The generated offspring then replace the least-fit parent classifiers within the population. A new generation of populations is generated once the offspring fully replaced parent classifiers in the population. This process of mating pool selection, fitness assessment, mating, and replacement is repeated over generations, progressively creating better classifiers until a completion criterion is met. After the best classifiers are outputted, post-GP analyses are carried out to compute gene occurrence in the classifiers as well as to predict on new unknown samples. (B) The representation of a GP tree structure for an exemplified classifier, Gene[A]/Gene[B] > 3. In general, a GP classifier is represented as a tree-based structure composed of the terminal set and function set. The terminal set, in tree terminology, is composed of leaves (nodes without branches) and may represent as genes or constants. The function set is a set of operators such as arithmetic operators (+, -, -, H) or Boolean operators (AND, OR, NOT), acting as the branch points in the tree, linking other functions or terminals. (C) The representation of a crossover operator of GP tree.