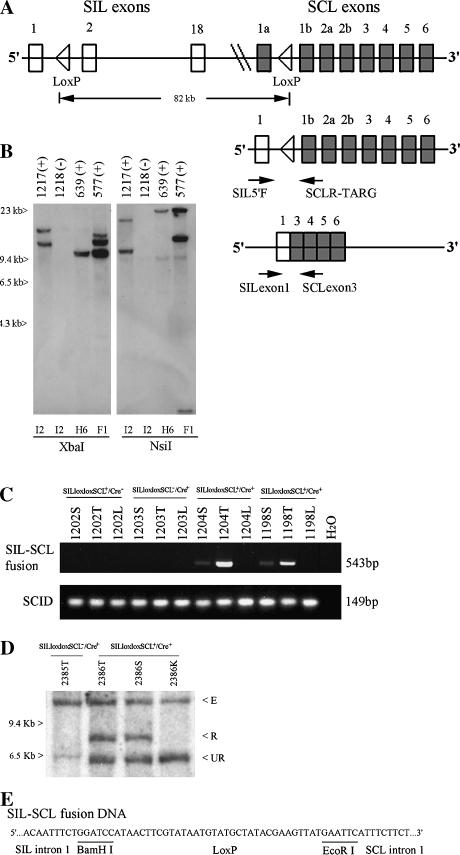
Figure 1.
Generation and characterization of SILloxloxSCL mice. (A) Top panel shows the human SIL (18 exons), SCL genes (8 exons), and the introduced Lox P sites in the regions of intron 1. Not all SIL exons are shown. Middle panel shows the fusion of genomic DNA. Two arrows indicate the primers used for a detection of fusion SILloxSCL DNA. Bottom panel shows the fusion mRNA that is generated between exon 1 of SIL and exon 3 of SCL in double-transgenic mice. Two arrows indicate the primers used for a detection of SIL-SCL fusion mRNA. (B) Detection of SILloxloxSCL BAC clone integration site(s) by Southern blot. Genomic DNA from F1 offspring of founders I2, H6, or F1 was digested with XbaI or NsiI and hybridized to a probe from the SCL 3′ untranslated region located at one terminal of the BAC clone. Variable-sized fragments representing unique integration sites can be seen. A cross-hybridizing band is seen in the NsiI digest at 9.4 kb. Mouse numbers are indicated, as is transgene status (+ or -) (C) SIL-SCL genomic fusion can be detected in mice 1204 and 1198 by PCR. Mice 1202, 1203, and H2O were negative controls. PCR amplification of the scid locus was used as a DNA quality control. T, thymus; S, spleen; L, liver. (D) Detection of SIL-SCL fusion by Southern blot analysis of SstI-digested genomic DNA. 2385T, thymus from mouse without the SILloxloxSCL transgene. 2386T, 2386S, 2386K; thymus, spleen, and kidney from SILloxloxSCL+/Cre+ mouse. E, endogenous murine SIL; R, recombined SIL-SCL fusion; UR, unrearranged SILloxloxSCL transgene. (E) Nucleotide sequences of fusion SILloxSCL genomic DNA.