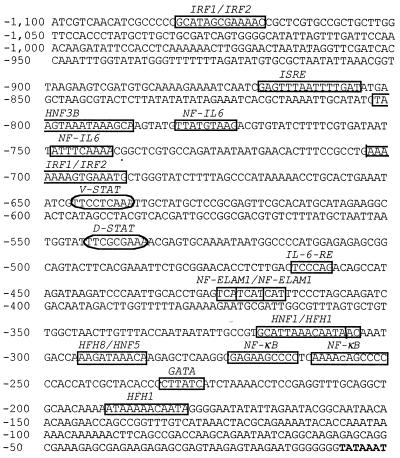
Figure 5.
Thor upstream region. Regulatory motifs are outlined in the sequence 5′ of the TATA box (bold). Consensus sequences used to identify these motifs are as follows, and referenced in 14, except as indicated: NFκB response elements, GGGRNTYYYY (31); GATA, WGATAR; IL-6 response elements (NF-IL6), TKNNGNAAK, (IL-6-RE) CTGGGA; Nuclear factor endothelial leukocyte adhesion molecule 1 (NF-Elam1), WCAKCAK; Hepatic nuclear factor 5 response element (HNF5), TRTTTGY. A single mismatch in one of the NFκB elements is indicated by a lowercase letter. Additional sequences with core identity and at least a 0.8 outside core match to the transfac database (20) also are indicated. These Transfac sequences are the elements for: interferon regulatory factors IRF1, SNAAAGYGAAACC, and IRF2, GNAAAGYGAAASY; interferon-stimulated response element IRSE, CAGTTTCWCTTTYCC; signal transducers and activators of transcription, Drosophila STAT (D-STAT), TTTCCSGGAAA and vertebrate STAT (V-STAT), TTCCCRKAA; hepatic nuclear factor HNF1, GGTTAATNWTTAMMN; hepatocyte nuclear factor/forkhead homolog HNF3B, KGNANTRTTTRYTTW, HFH1, NAWTGTTTATWT and HFH8, TGTTTATNYR. The core sequences are underlined. Only the GATA near NFκB is indicated.