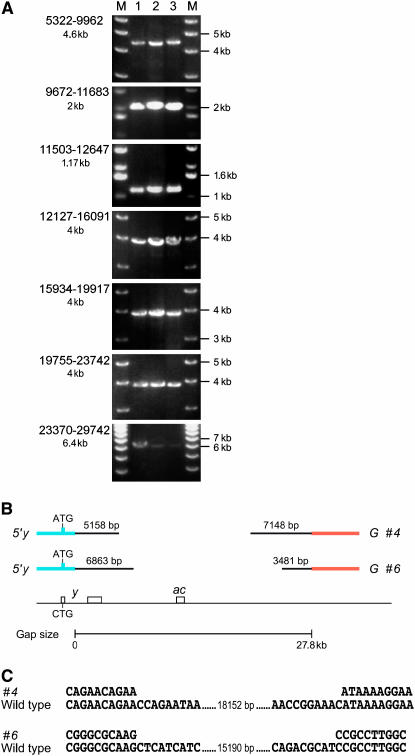
Figure 2.—
Examples of complete and partial gap repair events derived from the construct with a 28-kb gap (G). (A) PCR of three (lanes 1–3) complete events in the Df(1)y3PLsc8R background. The overlapping PCR products span a total of 24.4 kb corresponding to the downstream region of y. The primer pairs used are indicated on the left (see also Figure 1C). The size of the PCR product is indicated below the primer numbers. M, size marker. Primer numbers indicate the position relative to and downstream of the y transcription start. (B) Examples of two incomplete gap repair events (“#4” and “#6”). The in vivo-cloned DNA is in black with the size indicated above each segment. Orange and blue bars represent segments of the starting construct. The chromosomal region is shown below. Note that a considerable amount of DNA has been integrated at both ends of the DSB, but a gap of >15 kb remains. (C) Alignment of the DNA sequence at the junction of the events 4 and 6 with wild-type genomic DNA. The number indicates the length of the wild-type sequence not shown. Note the absence of (micro-)homology at the junction point.