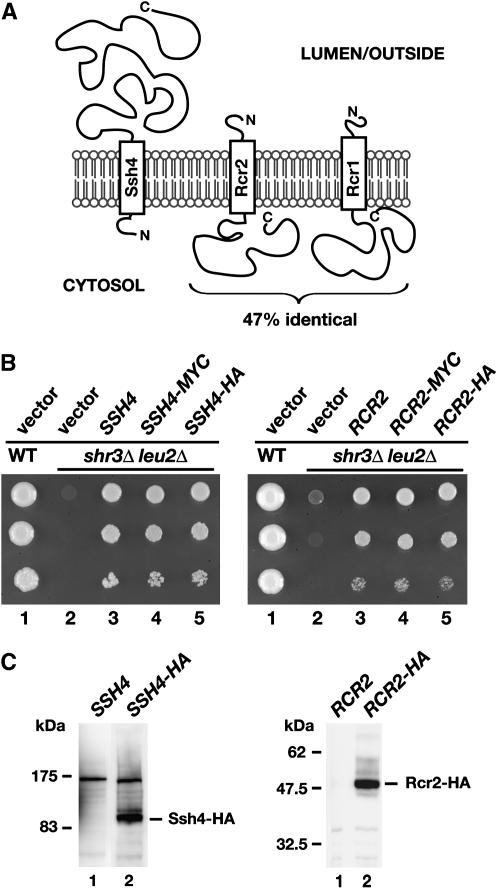
Figure 4.—
SSH4, RCR2, and RCR1 encode membrane proteins. (A) Schematic of secondary structure predictions of Ssh4, Rcr2, and Rcr1. Hydropathy analysis using a window size of 11 amino acid residues indicates that all three proteins harbor a single membrane-spanning segment in close proximity to their N termini (Kyte and Doolittle 1982). The orientation in the membrane is based on the N-best algorithm (TMHMM 1.0 prediction). (B) Functional analysis of epitope-tagged SSH4 and RCR2. Serial dilutions of strain HFY501 (WT: leu2 SHR3) transformed with pRS202 (vector) and strain HFY500 (leu2 shr3Δ) transformed with 2μ plasmids pRS202 (vector), pJK120 (SSH4), pJK131 (SSH4-3xMYC), pJK129 (SSH4-6xHA), pJK121 (RCR2), pJK135 (RCR2-3xMYC), or pJK133 (RCR2-6xHA) were applied to SC (−ura) plates. Plates were incubated at 30° for 3 days and photographed. (C) Immunoblot analysis of whole-cell extracts from strain JKY40 (ssh4Δ) transformed with either pJK120 (SSH4) or pJK130 (SSH4-6xHA) and strain JKY41 (rcr2Δ) transformed with either pJK121 (RCR2) or pJK134 (RCR2-6xHA), grown to log phase in SC (−ura) medium. Antibodies used to visualize Ssh4-HA and Rcr2-HA were α-HA (12CA5, mouse) and α-HA (3E10, rat), respectively.