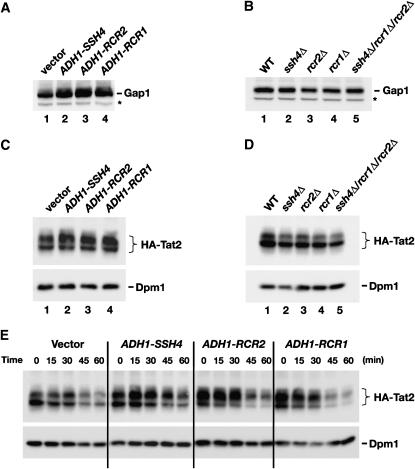
Figure 6.—
Analysis of Gap1 and Tat2 protein levels. (A) Gap1 levels in whole-cell extracts from a wild-type strain (CAY28) transformed with pRS202 (vector), pJK123 (PADH1-SSH4), pJK124 (PADH1-RCR2), or pJK125 (PADH1-RCR1). Cells were grown to an OD600 of 1.5 in SAD, and proteins in cell lysates were resolved by SDS–PAGE and immunoblotted with α-Gap1 antibodies. The asterisk indicates a nonspecific immunoreactive band unrelated to Gap1 that serves as a loading control. (B) Gap1 levels in whole-cell extracts from wild-type (CAY28), ssh4Δ (JKY40), rcr2Δ (JKY41), rcr1Δ (JKY42), and ssh4Δ rcr2Δ rcr1Δ (HFY538) strains. Cells were grown as in SAD supplemented with uracil, and extracts were prepared and analyzed as in A. (C) Steady-state levels of HA-TAT2 were analyzed in strain PLY860 cotransformed with pJK139 (HA-TAT2) and 2μ plasmid pRS202 (vector), pJK123 (PADH1-SSH4), pJK124 (PADH1-RCR2), or pJK125 (PADH1-RCR1) grown in SC (−ura,−trp) to an OD600 of 2. Cell extracts were resolved by SDS–PAGE and immunoblotted with α-HA (3F10, rat) and α-Dpm1 antibodies. (D) Immunoblotting of extracts from wild-type strain (CAY28), ssh4Δ (JKY40), rcr2Δ (JKY41), rcr1Δ (JKY42), and ssh4Δ rcr2Δ rcr1Δ (HFY538) transformed with plasmid pAS55 (HA-TAT2). Cells were grown SC (−ura) to an OD600 of 2 and proteins were resolved and analyzed as in C. (E) Strain PLY860 cotransformed with plasmids as in C was grown in SC (−ura −trp) to an OD600 of 2. Rapamycin was added and whole-cell extracts were prepared at the time points indicated. Proteins were resolved and analyzed as in C.