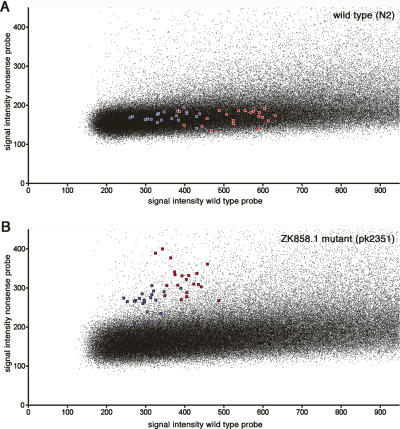
Figure 5.
Array-based NIM analysis. The signal of the nonsense (vertical) versus the wild-type (horizontal) allele is plotted for wild-type worms (N2) (A) and pk2351, containing an EMS-induced stopcodon in ZK858.1 (B). For nonmutated positions, the nonsense signal (vertical axis) is expected to be background, whereas the wild-type signal spreads over the X-axis, most likely reflecting GC content differences. A heterozygous mutation would have similar intensities for both the nonsense and wild-type allele, and these are thus expected to be located close to the diagonal in the figure. Microarrays with 390,000 features were used to assay part of the C. elegans genome-wide NIM collection. Every NIM was represented by four probes that were temperature-normalized (Tm = 72°C, probe length between 25 and 40 nt), representing the wild-type and mutant alleles for both DNA strands. The stop codon in ZK858.1 was represented 10 times (red, plus strand probes; blue, minus strand probes).