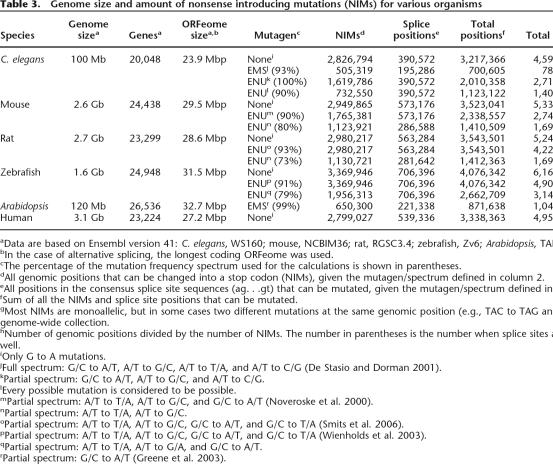
Table 3.
Genome size and amount of nonsense introducing mutations (NIMs) for various organisms
aData are based on Ensembl version 41: C. elegans, WS160; mouse, NCBIM36; rat, RGSC3.4; zebrafish, Zv6; Arabidopsis, TAIR6.0; human, NCBI36.
bIn the case of alternative splicing, the longest coding ORFeome was used.
cThe percentage of the mutation frequency spectrum used for the calculations is shown in parentheses.
dAll genomic positions that can be changed into a stop codon (NIMs), given the mutagen/spectrum defined in column 2.
eAll positions in the consensus splice site sequences (ag. . .gt) that can be mutated, given the mutagen/spectrum defined in column 2.
fSum of all the NIMs and splice site positions that can be mutated.
gMost NIMs are monoallelic, but in some cases two different mutations at the same genomic position (e.g., TAC to TAG and TAA) contribute to the genome-wide collection.
hNumber of genomic positions divided by the number of NIMs. The number in parentheses is the number when splice sites are taken into account as well.
iOnly G to A mutations.
jFull spectrum: G/C to A/T, A/T to G/C, A/T to T/A, and A/T to C/G (De Stasio and Dorman 2001).
kPartial spectrum: G/C to A/T, A/T to G/C, and A/T to C/G.
lEvery possible mutation is considered to be possible.
mPartial spectrum: A/T to T/A, A/T to G/C, and G/C to A/T (Noveroske et al. 2000).
nPartial spectrum: A/T to T/A, A/T to G/C.
oPartial spectrum: A/T to T/A, A/T to G/C, G/C to A/T, and G/C to T/A (Smits et al. 2006).
pPartial spectrum: A/T to T/A, A/T to G/C, G/C to A/T, and G/C to T/A (Wienholds et al. 2003).
qPartial spectrum: A/T to T/A, A/T to G/A, and G/C to A/T.
rPartial spectrum: G/C to A/T (Greene et al. 2003).